I'm trying to fit a GLM on my dataset which consists of soil respiration data (RS), soil temperature (TEMP), soil water content (SWC), biomass (BIOM), day of the year when the sampling was done (DOY) and the vegetation type (grasslands, old fields, ploughland and oversewn grassland). The measurement was done along a 15 m long circular transsect of consecutive quadrats, in every 20 cm, so there are 75 measurements in a transsect.
The question is the relationship between soil respiration (RS) and the other variables (SWC, TEMP, BIOM, DOY and type of vegetation) so how the changes of the related variables influence soil respiration (e.g. if the temperature is increasing, will soil respiration also increase?). I am thinking about a model like this: glm(RS~SWC+TEMP+DOY+type).
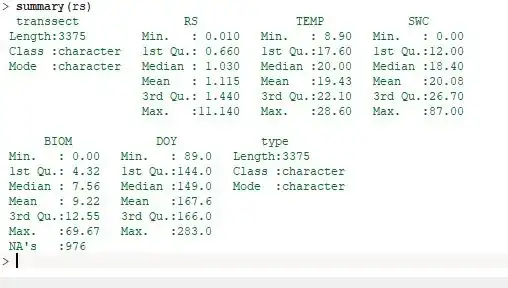
The values of RS, TEMP and DOY are all above zero, but SWC and BIOM have zero values, and there are NAs in the BIOM variable. None of the variables are normally distributed and there is an order of magnitude difference between the variables.
How can I decide which family to use?
Thank you for the suggestions!
Edit: boxplot and histogram of the variables
Related question: Do I need to transform my variables for GLM?




glm()function in the native stats package in R. ... That being said, depending on your audience, you might try a simple log transformation of RS instead. – Sal Mangiafico Nov 24 '22 at 17:29