The partial log-likelihood function in Cox proportional hazards is given with such formula $${}_{p}\ell(\beta) = \sum\limits_{i=1}^{K}X_i'\beta - \sum\limits_{i=1}^{K}\log\Big(\sum\limits_{l\in \mathscr{R}(t_i)}^{}e^{X_l'\beta}\Big),$$ where $K$ is the number of observations for which we have observed an event (where generaly there were $n$ observations, so $K-n$ observations were censored) and $\mathscr{R}(t_i)$ is a risk set for time $t_i$ defined as: $\mathscr{R}(t_i) = : \{X_j: t_j >= t_i, j = 1, \dots, n \}$.
I am trying to implement function calculating this partial log-likelihood function for given $\beta$ vector and input data set. I thought it's clever to first sort data by the observed time such that higher row number indicates higher survival time. For such a form of data I have prepared an implementation for 2 explanatory variables:
full_cox_loglik <- function(beta1, beta2, x1, x2, censored){
sum(rev(censored)*(beta1*rev(x1) + beta2*rev(x2) -
log(cumsum(exp(beta1*rev(x1) + beta2*rev(x2))))))
}
where beta1 is a coefficient for x1 variable, beta2 is a coefficient for x2 variable and censored is a vector indicating whether the observation had event (then $1$) or was censored (then $0$).
Being careful I have also prepared a second longer implementation that generally works for every dimension - it also takes data in a sorted format. This assumes that dCox is a data frame with explanatory variables and with censored column, beta is a vector of coefficients and status_number indicates the number of a column that has the information about censroing:
library(foreach)
partial_coxph_loglik <- function (dCox, beta, status_number) {
n <- nrow(dCox)
foreach(i=1:n) %dopar% {
sum(dCox[i,status_number]*(dCox[i,-status_number]*beta))
} %>% unlist -> part1
foreach(i=1:n) %dopar% {
exp(sum(dCox[i,-status_number]*beta))
} %>% unlist -> part2
foreach(i=1:n) %dopar% {
part1[i] - dCox[i, status_number]*(log(sum(part2[i:n])))
} %>% unlist -> part3
sum(part3)
}
Where to be more readable the partial log-likelihood would be (for that implementation):
$${}_{p}\ell(\beta) = \sum\limits_{i=1}^{K} \text{part1}_i - \sum\limits_{i=1}^{K}\log\Big(\text{part2}_i\Big).$$
For this implementation I have tried to calculate the values of the partial log-likelihood for the Cox proportional models for data that were generated from real $\beta$ parameters that were set to beta=c(2,2).
But I have received results telling me that the maximum of partial log-likelihood is not in the point beta=c(2,2) but far from this point.
One can prepare simulated survival data for Cox proportional hazards model, that came from Weibull distribution, with the metod explained here https://stats.stackexchange.com/a/135129/49793 . The similiar implementation based on that solution is below (works for 2 explanatory variables):
set.seed(456)
dataCox <- function(N, lambda, rho, x, beta, censRate){
# real Weibull times
u <- runif(N)
Treal <- (- log(u) / (lambda * exp(x %*% beta)))^(1 / rho)
# censoring times
Censoring <- rexp(N, censRate)
# follow-up times and event indicators
time <- pmin(Treal, Censoring)
status <- as.numeric(Treal <= Censoring)
# data set
data.frame(id=1:N, time=time, status=status, x=x)
}
x <- matrix(sample(0:1, size = 2000, replace = TRUE), ncol = 2)
dataCox(10^3, lambda = 5, rho = 1.5, x, beta = c(2,2), censRate = 0.2) -> dCox
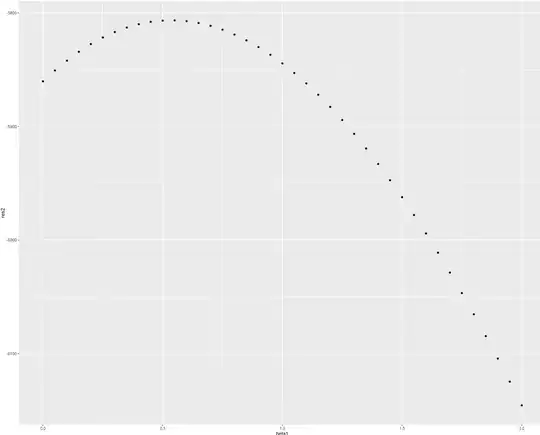
So for this implementation and simulated data I have calculated the values of partial log-likelihood for beta from c(0,0) to c(2,2) and received such results:
library(dplyr)
dCox %>%
dplyr::arrange(time) -> dCoxArr
beta1 <- seq(0,2,0.05)
beta2 <- seq(0,2,0.05)
res <- numeric(length(beta1))
for(i in 1:length(beta1)){
full_cox_loglik(beta1[i], beta2[i], dCoxArr$x.1,
dCoxArr$x.2, dCoxArr$status ) -> res[i]
}
library(ggplot2)
qplot(beta1, res)
res2 <- numeric(length(beta1))
for(i in 1:length(beta1)){
partial_coxph_loglik(dCoxArr[, c(4,5,3)],
c(beta1[i],beta2[i]), 3 ) -> res2[i]
cat("\r", i, "\r")
}
qplot(beta1, res2)
It does not look like the maximum of partial log-likelihood function is in the point beta = c(2,2) from which I have generated data. So now there appears my questio? Where did I make mistake? In data generation? In partial log-likelihood implementation? Or somewhere esle?


partial_log_likelihoodfunction looks pretty sloppy. Have you tried writing it in a sample sapply loop instead? One thing I notice in your derivation is that you use both failure and censoring times to index risk sets. Only the failure times index risk sets. If a individual is censored between two failure times, the arbitrary baseline hazard means you can't conclude anything extra except that they lived up to the most recent failure before they were censored. – AdamO Dec 17 '15 at 23:47One thing I notice in your derivation is that you use both failure and censoring times to index risk sets. Only the failure times index risk sets- but since the sum is over $\sum_{i=1}^{K}$ where the $K$ is a number of observed events I only add value to partial log-lik for observations with events. For censored observations I just multiple those parts by zero, which can be seen in receivingpart3:part1[i] - dCox[i, status_number]*(log(sum(part2[i:n]))). – Marcin Dec 17 '15 at 23:59survival::coxphfor each iteration step? – Marcin Dec 18 '15 at 00:15iter.max=1in the control and inputting the 1-step beta estimators as arguments toinitas I did before. – AdamO Dec 18 '15 at 17:17