Before I start, let me note that I have 0 experience with signal processing, so please bear with me:
My System
My system can be represented as an $m \times n$ matrix $X$ (input) where each column represents a "channel" and each row represents "time". A given input $x_{i,j}$ (time $i$ and channel $j$) produces:
- A signal $S = [R_0, R_1, R_2, ...]$ on channel $j$ starting at time $i$.
- A smaller (and inverted) signal on the neighboring channels. The scale by which the signal is reduced can be represented as a vector $B = [1, -\alpha_1, -\alpha_2, ... -\alpha_k, ...]$, where $k$ is the distance from the original channel $j$.
Lets assume that $S = [R_0, R_1, R_2]$ and $B = [1, -\alpha]$. Visually, this would look like: \begin{equation*} X = \begin{bmatrix} 0 & 1 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 \\ \vdots & \vdots & \vdots & \vdots & \vdots \\ 2 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 5 & 0 \\ 0 & 0 & 0 & 0 & 0 \\ \end{bmatrix} \implies Y = \begin{bmatrix} -\alpha R_0 & R_0 & -\alpha R_0 & 0 & 0 \\ -\alpha R_1 & R_1 & -\alpha R_1 & 0 & 0 \\ -\alpha R_2 & R_2 & -\alpha R_2 & 0 & 0 \\ \vdots & \vdots & \vdots & \vdots & \vdots \\ 2R_0 & -2\alpha R_0 & 0 & 0 & 0 \\ 2R_1 & -2\alpha R_1 & 0 & 0 & 0 \\ 2R_2 & -2\alpha R_2 & -5\alpha R_0 & 5R_0 & -5\alpha R_0 \\ 0 & 0 & -5\alpha R_0 & 5R_1 & -5\alpha R_1 \\ \end{bmatrix} \end{equation*}
I noticed that I can express this as \begin{equation*} Y = R X A \end{equation*}
where $R$ is a lower triangular matrix with $R_i$ on the $i$th diagonal: \begin{equation*} R = \begin{bmatrix} R_0 & 0 & 0 & \dots & \dots & 0 \\ R_1 & R_0 & 0 & \ddots & & \vdots \\ R_2 & R_1 & \ddots & \ddots & \ddots & \vdots \\ \vdots & \ddots & \ddots & \ddots & 0 & 0 \\ \vdots & & \ddots & R_1 & R_0 & 0 \\ R_{m-1} & \dots & \dots & R_2 & R_1 & R_0 \end{bmatrix} \end{equation*}
and $A$ is a symmetric matrix with $\alpha_i$ on the $i$th diagonal: \begin{equation*} A = \begin{bmatrix} 1 & -\alpha_1 & -\alpha_2 & \dots & \dots & 0 \\ -\alpha_1 & 1 & -\alpha_1 & \ddots & & \vdots \\ -\alpha_2 & -\alpha_1 & \ddots & \ddots & \ddots & \vdots \\ \vdots & \ddots & \ddots & \ddots & -\alpha_1 & -\alpha_2 \\ \vdots & & \ddots & -\alpha_1 & 1 & -\alpha_1 \\ 0 & \dots & \dots & -\alpha_2 & -\alpha_1 & 1 \end{bmatrix} \end{equation*}
My Current Solution
Given the nature of the $R$ matrix, any noise makes the solution blow up.
I am then setting my system as \begin{equation*} YA^{-1} = RX \end{equation*} And finding $X >= 0$ that minimizes \begin{equation*} \left| YA^{-1} - RX \right|_2 \end{equation*}
This works great. I am using Landweber iteration to solve this, and my solution looks good.
The problem is that it is too slow for my needs. I need to solve these systems in a hot loop millions of times.
My Actual Question
Do you have any suggestions?
I am thinking that I could treat every channel independently. The signals induced from another channel will just appear in this channel as a negative input (given that they have the same shape).
I can solve each single channel by expressing the convolution as the product of the FFTs. Then somehow try to match positive and negative signals in neighboring channels. Hopefully this "matching" could help mitigate random peaks from noise.
Can people suggest some reading materials where others solve similar problems? I don't know the technical terms, so I haven't had much success googling.
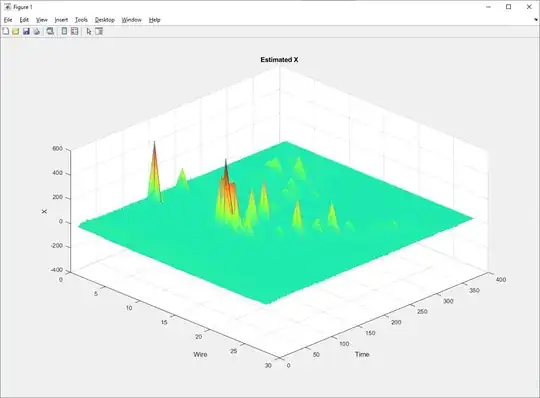
Some examples of deconvolved individual channels:
I get some ugly noise peaks that are comparable to actual signal peaks.


Edit 1
I have created a small git repository with some data and my sample solution using Landweber iteration. To reproduce it you can do:
Run:
git clone https://github.com/DJDuque/SP_Q.git
That will create the folder SP_Q.
Then go into the folder and run the Landweber iteration solution:
cd SP_Q
python landweber.py signal_*
That should produce a plot like:

Edit 2
I have also added the relevant stuff in CSV format:
- Y (observed signals): https://github.com/DJDuque/SP_Q/blob/main/signals.csv
- R matrix: https://github.com/DJDuque/SP_Q/blob/main/R.csv
- A matrix: https://github.com/DJDuque/SP_Q/blob/main/A.csv
- A_inv: https://github.com/DJDuque/SP_Q/blob/main/A_inv.csv
- Y * A_inv: https://github.com/DJDuque/SP_Q/blob/main/y_times_a_inv.csv









MATfile or something? – Royi Jul 01 '23 at 09:20npzfile orcsv? – Royi Jul 01 '23 at 14:30R,YandAfrom it? – Royi Jul 01 '23 at 14:41response.datin case you want to try the FFT method. – Daniel Duque Jul 01 '23 at 14:54