Short answer: cDNA is double-stranded.
I'll try to explain the concepts surrounding this with a nanopore read, where the cDNA component of the read (from the mouse genome) is indicated in a light grey, and the adapter components are either coloured or dark grey:
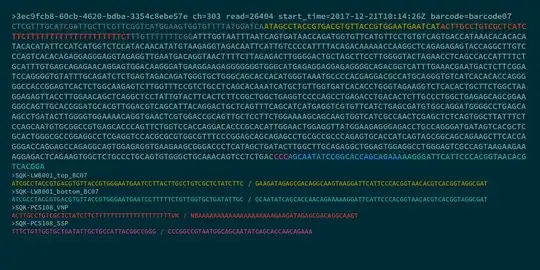
The cDNA library from this RNA was prepared by adding a polyT adapter (binding to the polyA sequence of the RNA), and carrying out reverse transcription to the start of the transcript. A strand-switching process was then used to prime and extend along the opposite strand of the DNA, creating a double-stranded full-length cDNA template. This template was then amplified using PCR to increase cDNA concentration and add in barcoding sequences.
You may notice that this particular read has a polyT head, rather than a polyA tail, attached to the VNP primer in the forward orientation. I can tell by this feature that this read comes from the reverse-complement strand of a DNA transcript, and so I know that the actual RNA transcript sequence is the reverse-complement of this read. Both the forward and reverse-complement sequences could potentially come through in the sequencer, because the adapters for sequencing are attached to both sides of the cDNA template, but the transcript direction can still be determined by looking at the orientation of the adapter sequences. When mapping this sequence to the genome, only a single strand (and its associated set of kmers) needs to be considered.
When considering what happens after mapping to the genome, there are four possible match situations:
- Adapter is in reverse orientation, genome is in reverse orientation
- Adapter is in forward orientation, genome is in reverse orientation
- Adapter is in reverse orientation, genome is in forward orientation
- Adapter is in forward orientation, genome is in forward orientation
In the case of the library preparation above, situations 2 and 3 correspond to a transcript that is encoded on the forward strand of the genome, whereas situations 1 and 4 correspond to a reverse-encoded transcript. Different library preparation methods could have a different way of encoding transcript direction. See here for more details on how these match situations can be used to produce strand-specific BAM files.
If I were to use an unstranded protocol (e.g. generating a cDNA library through other means, and attaching adapters randomly to the forward or reverse transcript strand), then the adapter direction wouldn't give me information about which strand this sequence came from. I would need to consider both strands when mapping (or more correctly, when identifying the native direction of the originating transcript), because the adapter sequence has been decoupled from the transcript direction.
On Illumina systems, this strand direction calculation is performed via the chemistry on the flow cell, and the adapters are [usually] not sequenced by the sequencer at all. Adapters from one direction bind to complementary sequences on the flow cell and are extended to produce the first read, and adapters from the other direction bind to produce the second read. Even with a stranded protocol, adapters from both directions are bound (because the cDNA template is double-stranded), but the binding order (as first or second read) can be used to determine the native direction of the originating transcript.
