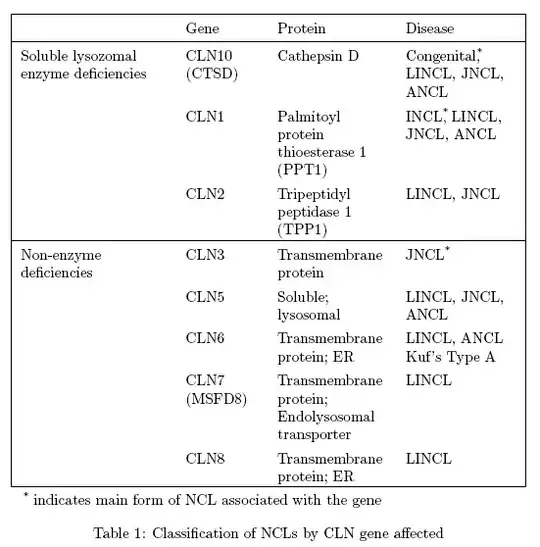
I'm getting really frustrated. I want to enter the following table into my LaTeX document, but the first and third columns refuse to narrow so it is too wide. Can someone please tell me what I am doing wrong? If I can format it properly the table should fit to about half a page width. Thanks for any help.
\begin{table}
\begin{tabular}{|c p{3cm}|c p{2cm}|c p{5cm}|c p{2cm}|}
\hline
& Gene & Protein & Disease \\
\hline
\multirow{3}{*}{Soluble lysozomal enzyme deficiencies} & CLN10 (CTSD) & Cathepsin D & Congenital*, LINCL, JNCL, ANCL \\
& CLN1 & Palmitoyl protein thioesterase 1 (PPT1) & INCL*, LINCL, JNCL, ANCL \\
& CLN2 & Tripeptidyl peptidase 1 (TPP1) & LINCL, JNCL \\
\multirow{5}{*}{Non-enzyme deficiencies} & CLN3 & Transmembrane protein & JNCL* \\
& CLN5 & Soluble; lysosomal & LINCL, JNCL, ANCL \\
& CLN6 & Transmembrane protein; ER & LINCL, ANCL Kuf's Type A \\
& CLN7 (MSFD8) & Transmembrane protein; Endolysosomal transporter & LINCL \\
& CLN8 & Transmembrane protein; ER & LINCL \\
\hline
\end{tabular}
\caption{Classification of NCLs by CLN gene affected \cite{haltia} \\ *indicates main form of NCL associated with the gene}
\label{table1}
\end{table}


\multirow; I'd say that shifting down the entries in the first column of this table hinders its readability, rather than enhancing it. – egreg May 31 '12 at 10:03\multirowor possibly an alternative means of row spanning would in my opinion be adequate here if the first-column entries were sufficiently long to determine the first row's height. – dgs May 31 '12 at 10:27