Hi I am using Cox regression to do some survival analysis. I have one continous predictor variable called "biomarker" and two binary categorical variables sex and diabetes.
I want to get the interaction p-values between sex and "biomarker", in both non-diabetics and diabetic patients.
I am currently using the following code:
coxph(
Surv(days2death, death) ~
factor(diabetes) :
(factor(male) * biomarker) +
factor(diabetes), data = df.cox.testing).
Am I correct in thinking that the last two rows correspond to interaction p-values between biomarker and sex at different levels of diabetes?
coef exp(coef) se(coef) z Pr(>|z|)
factor(diabetes)1 -1.682469 0.185914 1.116110 -1.507 0.132
factor(diabetes)0:factor(male)1 -0.168605 0.844842 0.879000 -0.192 0.848
factor(diabetes)1:factor(male)1 -0.271691 0.762089 1.013834 -0.268 0.789
factor(diabetes)0:biomarker 0.108440 1.114538 0.130682 0.830 0.407
factor(diabetes)1:biomarker 0.224785 1.252054 0.145329 1.547 0.122
factor(diabetes)0:factor(male)1:biomarker -0.045544 0.955477 0.168073 -0.271 0.786
factor(diabetes)1:factor(male)1:biomarker -0.008174 0.991859 0.166480 -0.049 0.961
I created a forest plot with the results. However the overlap of the confidence intervals doesnt seem to fit visually with the extracted p-values. (Blue is female, red is male)
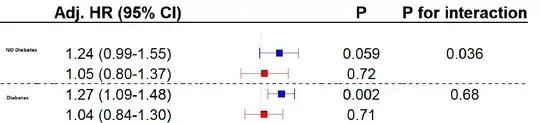
maleas a predictor in its own right, outside of the interaction? It can be confusing to interpret interactions when one of the included predictors doesn't have its own term in the model. – EdM Mar 19 '24 at 15:20maleit does not change anything about the model/output/p-values. But I can see how that can be confusing. – Fabian Mar 19 '24 at 15:27emmeanspackage, or those built in to thermspackage for its own functions, to make any comparisons or display that you want from the correct model. – EdM Mar 19 '24 at 15:35