I want to analyze forest regeneration after a forest fire. I have a data frame containing NDVI values (NDVI_mean) (NDVI is a vegetation index for plant vigor) of different study sites (Treatment) over time (date). The study sites are grouped in different management treatments (Management1). This is the head and tail of the data:
Treatment NDVI_mean date Management1
1 G 0.7918969 2018-10-11 Control
2 J 0.1899928 2018-10-11 Clearcut
3 H 0.2378630 2018-10-11 Clearcut
4 C 0.3631807 2018-10-11 PR1/2
5 D 0.3294372 2018-10-11 PR1/2
6 E 0.4611494 2018-10-11 PR3/4
7 F 0.3681659 2018-10-11 PR3/4
8 I 0.3020894 2018-10-11 Clearcut
9 B 0.3247002 2018-10-11 NR
10 K 0.3109387 2018-10-11 NR
11 L 0.8071204 2018-10-11 Control
12 G 0.8191044 2018-11-28 Control
13 J 0.1952865 2018-11-28 Clearcut
14 H 0.2209233 2018-11-28 Clearcut
15 C 0.3942993 2018-11-28 PR1/2
16 D 0.3285107 2018-11-28 PR1/2
17 E 0.4192142 2018-11-28 PR3/4
18 F 0.3856550 2018-11-28 PR3/4
19 I 0.2522038 2018-11-28 Clearcut
20 B 0.3300189 2018-11-28 NR
21 K 0.2990551 2018-11-28 NR
22 L 0.8361506 2018-11-28 Control
23 G 0.8274411 2018-12-05 Control
...
479 E 0.5348337 2022-06-15 PR3/4
480 F 0.4959483 2022-06-15 PR3/4
481 I 0.5295775 2022-06-15 Clearcut
482 B 0.4168112 2022-06-15 NR
483 K 0.4540260 2022-06-15 NR
484 L 0.6940871 2022-06-15 Control
I want to test if there are significant differences in NDVI_mean between the different groups (Management1). However, the results of the mixed-effect model I use in lme4 seem a bit off, as they indicate that there are no significant differences between any group except for the control group to others.
This is what I have tried so far:
model <- lmer(NDVI_mean ~ Management1 + (1 | Treatment), data = data_all)
summary(model)
mc <- glht(model, linfct = mcp(Management1 = "Tukey"))
summary(mc)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lmer(formula = NDVI_mean ~ Management1 + (1 | Treatment), data = data_all)
Linear Hypotheses:
Estimate Std. Error z value Pr(>|z|)
Control - Clearcut == 0 0.336027 0.029192 11.511 <1e-04 ***
NR - Clearcut == 0 -0.038349 0.029192 -1.314 0.682
PR1/2 - Clearcut == 0 -0.031177 0.029192 -1.068 0.822
PR3/4 - Clearcut == 0 0.019608 0.029192 0.672 0.962
NR - Control == 0 -0.374376 0.031979 -11.707 <1e-04 ***
PR1/2 - Control == 0 -0.367204 0.031979 -11.483 <1e-04 ***
PR3/4 - Control == 0 -0.316419 0.031979 -9.895 <1e-04 ***
PR1/2 - NR == 0 0.007173 0.031979 0.224 0.999
PR3/4 - NR == 0 0.057957 0.031979 1.812 0.365
PR3/4 - PR1/2 == 0 0.050785 0.031979 1.588 0.504
Signif. codes: 0 ‘*’ 0.001 ‘’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Adjusted p values reported -- single-step method)
Looking at the plotted data I would expect the differences of some of the groups to be significant:
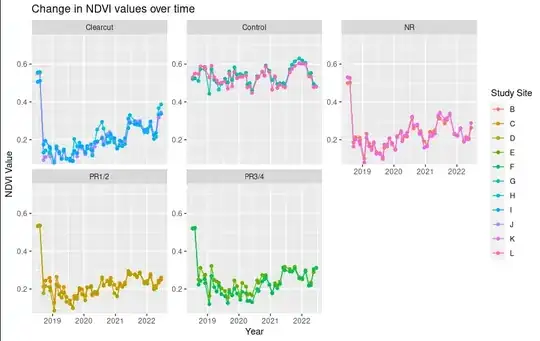
I am wondering if I am overseeing something with my code or there are just no significant differences.
EDIT
I should have clarified my research question better in the first place:
I want to analyze, which Management1 group has a stronger effect on NDVI_mean over time.
As a first step, I switched to lme as suggested by Shawn. As I want to evaluate the changes of NDVI_meanover time, this is my code:
model <- lme(NDVI_mean ~ Management1, random = ~1 | date, correlation = corAR1(form = ~ 1 | date), data = Data_2018_2022)
With the following output:
Linear mixed-effects model fit by REML
Data: Data_2018_2022
AIC BIC logLik
-1240.825 -1207.452 628.4127
Random effects:
Formula: ~1 | date
(Intercept) Residual
StdDev: 0.07154796 0.05647643
Correlation Structure: AR(1)
Formula: ~1 | date
Parameter estimate(s):
Phi
0.09700646
Fixed effects: NDVI_mean ~ Management1
Value Std.Error DF t-value p-value
(Intercept) 0.3193368 0.011910976 436 26.81030 0.0000
Management1Control 0.4485332 0.007679661 436 58.40534 0.0000
Management1NR 0.0307109 0.007887175 436 3.89377 0.0001
Management1PR1/2 0.0246634 0.007886995 436 3.12709 0.0019
Management1PR3/4 0.0580982 0.007886995 436 7.36633 0.0000
Correlation:
(Intr) Mngm1C Mng1NR M1PR1/
Management1Control -0.265
Management1NR -0.257 0.409
Management1PR1/2 -0.256 0.377 0.366
Management1PR3/4 -0.256 0.377 0.366 0.399
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-3.00833022 -0.53491680 -0.03658359 0.59397622 4.30824733
Number of Observations: 484
Number of Groups: 44
When I plot the normalized residuals plot(ACF(model, resType = "normalized"), alpha = 0.05)
The plot looks like this.

It seems like there is still an autocorrelation. Is my overall approach valid, or am I overseeing something?
EDIT2
I have followed the suggestions of @Edm and fit the model as following:
model <- gls(NDVI_mean ~ Management1 + splines::ns(year_month_numeric), correlation = corCAR1(form = ~ year_month_numeric | Treatment), data = Data_2018_2022)
I run into the problem that when I want to include an interaction of Management1 and date Data_2018_2022$Management1_date <- interaction(Data_2018_2022$Management1,Data_2018_2022$year_month_numeric) in the model model <- gls(NDVI_mean ~ Management1_date + splines::ns(year_month_numeric), correlation = corCAR1(form = ~ year_month_numeric | Treatment), data = Data_2018_2022), I get the error Error in glsEstimate(object, control = control) : computed "gls" fit is singular, rank 221
Also, there is still a autocorrelation in the residuals:

NDVI_meanover time. Your plots suggest that, if there are any differences amongManagement1values, they are in those patterns over time:PR1/2is almost flat after the initial drop, whileClearcutseems to rise. Also, your analysis seems to include the initial (pre-intervention?) outcome values, before the big drops, along with all the later values, muddying the comparisons. Look at Frank Harrell's chapter on longitudinal responses, along with Shawn's answer and Robert's comments. – EdM Jan 11 '24 at 16:22dateand its interaction withManagement1, use standard linear regression with robust standard errors instead of a correlation structure to account for the repeated measures. – EdM Jan 19 '24 at 13:59