I have a non linear regression model that is based on an exponential decay function. When I try to add a confidence interval, instead of using the models in built 'confidence interval' prediction functionality, I wanted to do so manually. I get my model params and their standard errors, then calculate the prediction using the param +/- 2 standard errors. The plot I work on in the code below shows my final result. Expectation was to have a upper and lower confidence interval line encompassing most of the data, but instead it looks like it just passes through the middle.
Example data:
library(tidyverse)
example_data <- dput(example_data)
structure(list(cohort_id = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 9L, 9L,
9L, 9L, 9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 10L,
10L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L,
11L, 11L, 11L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L,
12L, 12L), levels = c("1", "2", "3", "4", "5", "6", "7", "8",
"9", "10", "11", "12"), class = "factor"), billing_cycle = c(1L,
2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L,
5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L,
8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L,
4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L,
7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L,
10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L,
12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L), sample_survival_rate = c(0.630375321185327,
0.467011817658967, 0.365716219971609, 0.320696843452609, 0.289715829574683,
0.274138331734878, 0.263859709445232, 0.257975988868186, 0.25428803817773,
0.253349513689708, 0.251974683159173, 0.25087864807282, 0.626381321766971,
0.457700237294344, 0.37100891459414, 0.322040406927193, 0.294130414649021,
0.273224287881716, 0.264008524367158, 0.257890204475791, 0.254600100163038,
0.252665019457592, 0.250872931127164, 0.249283495816814, 0.617466244254771,
0.463682683415105, 0.371605156168376, 0.320962165438288, 0.294378622257423,
0.272094922902315, 0.263553461969235, 0.258151398805649, 0.254228898501473,
0.252721988646528, 0.250957888359671, 0.250678608369381, 0.625842284676012,
0.475715043015969, 0.364457416055596, 0.326485932788029, 0.287971100886182,
0.272805038087397, 0.263982745776158, 0.258071343467853, 0.254782618390382,
0.252538854336153, 0.251931200298009, 0.250189014687092, 0.614958361152235,
0.474023154666957, 0.37790296469348, 0.321305725376115, 0.288431205767636,
0.273448941036557, 0.265221683162653, 0.257977862090824, 0.254402493942438,
0.252262229858011, 0.252482170616736, 0.250167038793874, 0.615276997378503,
0.462251124297695, 0.379117488405097, 0.320048927623227, 0.286479327627075,
0.277112140101815, 0.264517062972436, 0.257970078541396, 0.254324910083164,
0.253199527200694, 0.250985688661134, 0.251435584163309, 0.626165857232067,
0.457716584851464, 0.380706922662529, 0.32214396867372, 0.28725348189702,
0.274217428106078, 0.262850951863544, 0.258008309979443, 0.253982997355457,
0.252432836066219, 0.252955446219195, 0.250173801962517, 0.626248558471049,
0.474358329360571, 0.369797644382662, 0.324484196652504, 0.292835709935179,
0.274812679385776, 0.264902105112049, 0.257920928121361, 0.254293689364316,
0.252094179645866, 0.25146035913424, 0.251382845450947, 0.619935506915612,
0.472368360759675, 0.376906826285281, 0.322622472724279, 0.294056956175115,
0.2762753843963, 0.263543866656661, 0.257911561154227, 0.254082172527871,
0.252479306436864, 0.25092022041681, 0.25147680046597, 0.620940410953731,
0.458684083447818, 0.367775959111921, 0.31955299117994, 0.290834501870578,
0.273073930174107, 0.264099158034426, 0.258049918137067, 0.254097282656084,
0.252593088721244, 0.251201515264958, 0.249725999038376, 0.6178781171174,
0.480537380689229, 0.370305282109615, 0.321910535895373, 0.295165049355668,
0.274898706563092, 0.264775605155201, 0.257970685836666, 0.254379583052875,
0.25230912983428, 0.251323575117219, 0.250613209856686, 0.627625590735999,
0.470453669107974, 0.361352411452518, 0.329163446065758, 0.293046687979046,
0.275398723193211, 0.263940477049976, 0.257914465199765, 0.254451201344154,
0.251946651930126, 0.251414402429264, 0.252082405269007)), row.names = c(NA,
-144L), class = c("tbl_df", "tbl", "data.frame"))
The above block creates a data frame example_data. Here's a plot of the fields of interest:
survival_plot <- example_data |>
ggplot(aes(x = billing_cycle, y = sample_survival_rate, color = cohort_id)) +
geom_line()
survival_plot
Then I fit a model. After fitting I retrieve the model params i, a and lambda along with their standard errors, then I (attempt to) add an upper and lower bound confidence interval:
# function
exponential_decay <- function(i, a, lambda, billing_cycle) i + a * exp(-lambda * billing_cycle)
mod.nls <- nls(sample_survival_rate ~
exponential_decay(i, a, lambda, billing_cycle), data = example_data, start = list(i = 0.5, a = 0.5, lambda = 0.15))
mod_summary <- mod.nls |> summary()
mod_i <- coef(mod.nls)['i']
mod_i_se <- mod_summary$coefficients["i", "Std. Error"]
mod_a <- coef(mod.nls)['a']
mod_a_se <- mod_summary$coefficients["a", "Std. Error"]
mod_lambda <- coef(mod.nls)['lambda']
mod_lambda_se <- mod_summary$coefficients["lambda", "Std. Error"]
add 95% CI to example data
example_data <- example_data |>
mutate(
mod_upper_ci = exponential_decay(mod_i + 2(mod_i_se), mod_a + 2(mod_a_se), mod_lambda + 2(mod_lambda_se), billing_cycle),
mod_lower_ci = exponential_decay(mod_i - 2(mod_i_se), mod_a - 2(mod_a_se), mod_lambda - 2(mod_lambda_se), billing_cycle)
)
But when I add the upper/lower interval to my plot I get this:
survival_plot +
geom_line(aes(x = billing_cycle, y = mod_upper_ci), color = 'black') +
geom_line(aes(x = billing_cycle, y = mod_lower_ci), color = 'black')
Does this look 'right'? I expected the black lines to encompass the cohort_ids, instead it looks like it's just going through the middle.
Is my approach flawed (presumably yes)? How can I correctly add a 95% confidence interval to my plot using the model params and their standard errors?

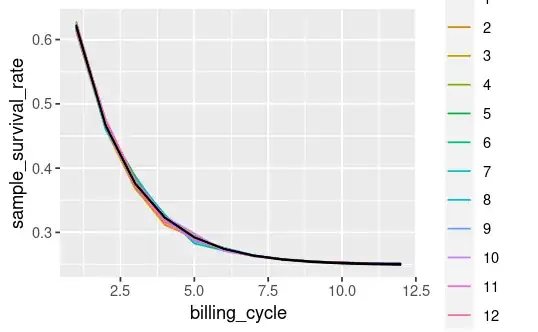
cohort_idsare basically your data points. The 95% CI is the CI on the mean. It isn't a prediction interval. The prediction interval should cover the data. The CI just covers the mean. So it's probably doing the right thing (I can't reproduce your modelling from the code given in the question) – Alex J Jul 12 '23 at 06:01exponential_decayfunction is defined – Alex J Jul 12 '23 at 06:36exponential_decay <- function(i, a, lambda, billing_cycle) i + a * exp(-lambda * billing_cycle)– Doug Fir Jul 12 '23 at 07:15