There are several ways to report the estimated outcomes from an ordinal regression model. With the emmeans package you can choose among them with a mode argument submitted to emmeans() or ref_grid().
Latent variable
According to a vignette page, the default is mode = "latent", and
With mode = "latent", the reference-grid predictions are made on the scale of the latent variable implied by the model. The scale and location of this latent variable are arbitrary...
The results you show are based on that underlying default "latent variable" model, an assumed continuous response (arbitrarily centered and scaled) that is converted to ordered categories as its value passes associated thresholds. The idea is illustrated in Figure 6.4 of these course notes. That "latent variable" model is fundamental and can be used to obtain other types of outcome estimates.
As you discovered, the "latent variable" for a scenario here is just the linear predictor $X' \beta$, where $X$ is the vector of predictor values and $\beta$ is the corresponding vector of coefficient estimates. When you ask emmeans for estimates of marginal means, it takes the grid of values of variables that you specify (in this case, the combinations of the predictors ses and fam) and returns the model's estimates for those values (by default, at the average values of variables that you don't specify).
As the vignette notes, however, "The scale and location of this latent variable are arbitrary." In the parameterization used by the lrm() and orm() functions in the rms package, the linear predictor is associated with the class probabilities:
$$\Pr(Y \ge j|X)=\frac{1}{1+\exp\left(-(\alpha_j + X' \beta)\right)} $$
where you have classes of outcome $Y$ labeled with increasing values of $j$ and the $\alpha_j$ are the corresponding intercepts estimated by the model. Clearly, you can shift the linear predictor $X' \beta$ by any offset and get the same outcome probability estimates if you make corresponding changes to the values of the $\alpha_j$ intercepts.
From that perspective, you can display the latent-variable estimates with any offset shift that you want. The mode="middle" choice will tend to center the estimates around 0, if you want to display the results in terms of the latent variable. It doesn't matter that the model estimates you displayed have negative values. If you ask for probability estimates, the corresponding values of $\alpha_j$ (your two intercept estimates) will put everything together properly.
Probability estimates
If you want outcome-class probabilities, you need to specify a different mode. There is a mode="prob" argument that can report class probability estimates for supported model types.
There's a "gotcha" with that mode, however, explained in the section of the vignette on multinomial responses, of which ordinal models are a special case:
Please note that, because the probabilities sum to 1 (and the latent values sum to 0) over the multivariate-response levels, all sensible results from emmeans() must involve that response as one of the factors. For example, if resp is a response with k levels, emmeans(model, ~ resp | trt) will yield the estimated multinomial distribution for each trt; but emmeans(model, ~ trt) will just yield the average probability of 1/k for each trt.
If you want to display class probabilities, you need to include the outcome among the variables that you specify to emmeans(). For example, to get a plot of outcome-class probabilities as a function of both ses and fam, you could specify:
emmip(int, outcome ~ fam|ses, mode = "prob", CIs = TRUE)
That will display estimates of outcome probabilities as a function of fam, in a separate facet for each ses level. The outcome probabilities for each combination of fam and ses will sum to 0, as expected.
A warning about implementation: in the older version of emmeans I'm using on this computer (emmeans_1.6.2-1), the mode="prob" argument didn't seem to work on an lrm() model, while it worked OK with an orm() model (which provided the same coefficient estimates). I don't know if that's still the case with newer versions.
Contrasts and statistical significance
A useful estimate of the "significance" of your predictors with respect to outcome includes all the terms involving each predictor. If you use a model generated by the rms package, the anova() function applied to the model provides a very useful display of overall significance and the significance of interaction and nonlinear terms in the model, via Wald tests.
At the other extreme of complexity, the pairwise comparisons among all predictor combinations take into account the corresponding coefficient estimates $\beta$, their variances, and their covariances. It applies the formula for the variance of a weighted sum of variables to determine if any of those coefficient combinations differs significantly from 0, with multiple-comparison corrections as specified.
Of course, the correction for multiple comparisons becomes more restrictive as the number of comparisons increases. If there are particular comparisons of major interest, it can be better to restrict your analysis to those rather than evaluating all pairwise comparisons.
For those pairwise differences, any constant offset shift in the linear predictors don't matter, as the offset will cancel out when you take the differences. That's another reason not to worry about the negative values displayed for the latent variables.
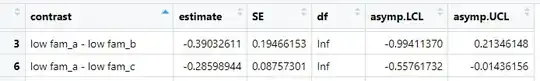
The point estimates of both of the contrasts that you show agree with the differences of the corresponding emmean values in the table you show. The standard error of low fam_a - low fam_b, however, is more than twice as large as for low fam_a - low fam_c, presumably because of a small number of cases in low fam_b. Although the point estimate of low fam_a - low fam_b is larger than for low fam_a - low fam_c, the larger standard error means that the first contrast isn't significant at $p < 0.05$ while the second is.
A final warning: overlapping confidence intervals can be consistent with a significant difference between two scenarios. The emmeans FAQ emphasizes that point. This answer goes into detail with respect to t-tests in simple scenarios, in which non-overlap of 95% CI is approximately equivalent to $p < 0.005$ for the difference between means.


polr()versuslrm()versusorm()) and the call you made toemmeans()to get the table of contrasts that you show. – EdM May 13 '23 at 17:02