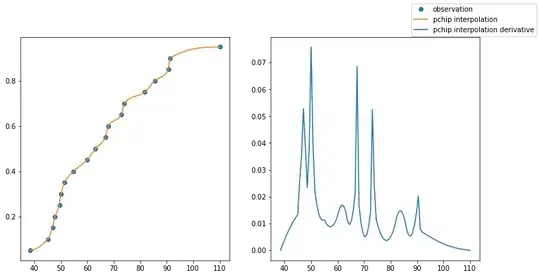
I would like to determine CDF and PDF from quantiles that I have determined via quantile regression. I have read here in the forum (Find the PDF from quantiles) that it is possible to interpolate this via the integral of a B-spline The PDF should then be determined via a normal evaluation. Unfortunately I did not understand why I have to use the integral of the B-spline, how can I ensure that the CDF is monotonically increasing and how I then get to the derivative (the PDF)? Can someone help me please?
This is how it currently looks for me:
import scipy.interpolate
import numpy as np
x = np.array([ 38.45442808, 45.12051933, 46.85565437, 47.84576924,
49.50084204, 50.09833301, 51.3717386 , 54.85307741,
59.91982266, 63.11786854, 66.90037244, 67.84446378,
72.96120777, 73.92993279, 81.63075081, 85.42178836,
90.70554533, 91.2393176 , 110.03872988])
y = np.array([0.05, 0.10, 0.15, 0.20, 0.25, 0.30, 0.35, 0.40, 0.45, 0.50, 0.55, 0.60, 0.65, 0.70, 0.75, 0.80, 0.85, 0.90, 0.95])
t,c,k = scipy.interpolate.splrep(x, y)
spline = scipy.interpolate.BSpline(t, c, k, extrapolate=False)
d_spline = spline_.derivative()
N = 100
xmin, xmax = x.min(), x.max()
xx = np.linspace(xmin, xmax, N)
fig, ax = plt.subplots(2,1, figsize =(12, 8))
ax[0].plot(x, y, 'bo', label='Original points')
ax[0].plot(xx, spline(xx), 'r', label='BSpline')
ax[1].plot(xx, d_spline(xx), 'c', label='BSpline')
My approach doesn't really work well unfortunately and I can't find any numerical examples to help me. I am grateful for all comments and remarks!
Thank you!