I am currently at a dilemma concerning a model describing the allometric relationship between body size and mass. After carefully checking model assumptions and selecting the model that best fits the data, my final model was the following :
modD=lmer(body_size~0+(D_Mass*Species*sex+D_Mass*sex*Season +Species*Season)+(1|site), data=Rhabdoglobal, REML= T)
Diagnostic plots showed no indication of non-linearity or violating the normality of residuals or heteroscedasticity of variances. 0 was chosen for the intercept as biologically we know for a fact that at 0 body size there is 0 mass, so the relationship between the two must always cross the point (0,0). These were the results of the Anova :
Type III Analysis of Variance Table with Satterthwaite's method
Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
D_Mass 214.78 214.782 1 676.03 717.7672 < 2.2e-16 ***
Species 510.73 255.365 2 120.41 853.3874 < 2.2e-16 ***
sex 0.95 0.950 1 678.15 3.1732 0.0753052 .
Saison 8.71 8.706 1 680.00 29.0926 9.533e-08 ***
D_Mass:Species 0.01 0.007 1 678.49 0.0231 0.8791882
D_Mass:sex 0.01 0.014 1 677.11 0.0456 0.8308968
Species:sex 0.96 0.964 1 677.12 3.2204 0.0731712 .
D_Mass:Saison 7.02 7.017 1 677.22 23.4497 1.590e-06 ***
sex:Saison 3.90 3.904 1 676.44 13.0453 0.0003265 ***
Species:Saison 0.26 0.257 1 562.45 0.8579 0.3547159
D_Mass:Species:sex 1.08 1.082 1 676.27 3.6148 0.0576921 .
D_Mass:sex:Saison 3.59 3.590 1 675.85 11.9973 0.0005664 ***
However, when I try to illustrate my results in plot form, the relationship does not seem linear at all.

Rather, changing the plot expression to y=log(x) seems to solve that problem :
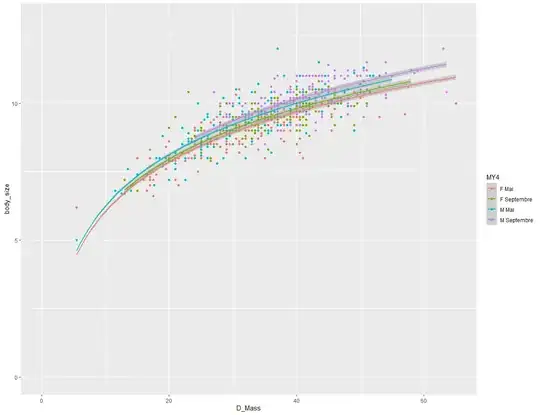
My questions would be the following :
- Is it possible to represent a relationship that was described by a statistical model using a different expression than if one would draw a plot directly from the model estimates ?
- If not, is it justifiable to use a different model, not necessarily better in terms of linearity, homoscedasticity or normality of residuals, but simply based on post-hoc representation of raw data ?
Edit : thank you for the response. Using a backward stepwise method,this is the simplest model with the lowest AIC that I came up with.