I have fit a GAM with two continuous independent variables and one discrete covariate with two levels ("gray" and "brown"). All variables are scaled and the model runs fine:
mod3 = gam(cont1 ~ disc1 * cont2 * cont3 +
s(cont2, cont3, by=disc1, k=4),
data=data_so)
summary(mod3)
Family: gaussian
Link function: identity
Formula:
cont1 ~ disc1 * cont2 * cont3 + s(cont2, cont3, by = disc1, k = 4)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.015009690 0.038191859 0.39301 0.69440639
disc1brown -0.032717084 0.066537035 -0.49171 0.62304187
cont2 0.212856916 0.018793283 11.32622 < 2.22e-16
cont3 0.005457858 0.019394264 0.28142 0.77845557
disc1brown:cont2 0.061748331 0.021911015 2.81814 0.00493549
disc1brown:cont3 0.016191669 0.027321828 0.59263 0.55357838
cont2:cont3 0.118947202 0.035662553 3.33535 0.00088654
disc1brown:cont2:cont3 -0.098224982 0.071036399 -1.38274 0.16708507
Approximate significance of smooth terms:
edf Ref.df F p-value
s(cont2,cont3):disc1gray 1.8362810 2.0676743 1.08907 0.1846056
s(cont2,cont3):disc1brown 0.7998747 0.7998747 9.65092 0.0055757
Rank: 10/14
R-sq.(adj) = 0.128 Deviance explained = 13.5%
GCV = 0.88013 Scale est. = 0.87182 n = 914
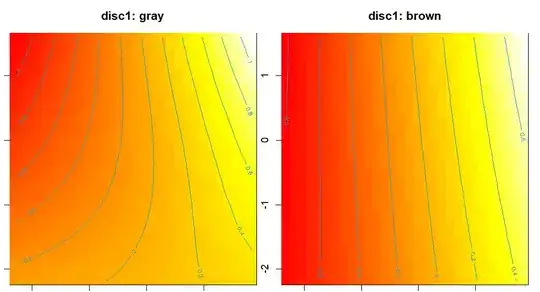
However, the approximate significance of each smooth term does not match my a priori expectation. More importantly, it also doesn't match the visualized model fit, by which "gray" should be non-linear, and "brown" somewhat linear:
vis.gam(mod3, view=c("cont2","cont3"), plot.type="contour",
cond=list(disc1="gray"), main="disc1: gray")
vis.gam(mod3, view=c("cont2","cont3"), plot.type="contour",
cond=list(disc1="brown"), main="disc1: brown")
If I run the model only with either class, it appears to match the figure (high F/low P value for approx. sign. of smooth term for "gray, and the opposite for the "brown" model:
> mod3a = gam(cont1 ~ cont2 * cont3 +
+ s(cont2, cont3, k=4),
+ data=data_so[disc1=="gray"])
> summary(mod3a)
Family: gaussian
Link function: identity
Formula:
cont1 ~ cont2 * cont3 + s(cont2, cont3, k = 4)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.021064013 0.038786204 0.54308 0.5872749
cont2 0.175710375 0.019469841 9.02475 < 2.22e-16
cont3 -0.003258934 0.019493737 -0.16718 0.8672855
cont2:cont3 0.118727033 0.036447026 3.25752 0.0011869
Approximate significance of smooth terms:
edf Ref.df F p-value
s(cont2,cont3) 1.590295 1.828967 34.76433 < 2.22e-16
Rank: 5/7
R-sq.(adj) = 0.127 Deviance explained = 13.2%
GCV = 0.91631 Scale est. = 0.90938 n = 609
> mod3b = gam(cont1 ~ cont2 * cont3 +
s(cont2, cont3, k=4),
data=data_so[disc1=="brown"])
> summary(mod3b)
Family: gaussian
Link function: identity
Formula:
cont1 ~ cont2 * cont3 + s(cont2, cont3, k = 4)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.01997134 0.05075102 -0.39352 0.69422
cont2 0.17893076 0.02588782 6.91177 0.000000000028642
cont3 0.01326603 0.02519297 0.52658 0.59888
cont2:cont3 0.02072222 0.05873067 0.35283 0.72446
Approximate significance of smooth terms:
edf Ref.df F p-value
s(cont2,cont3) 1.075582 1.075582 2.71988 0.06871
Rank: 5/7
R-sq.(adj) = 0.133 Deviance explained = 14.2%
GCV = 0.80732 Scale est. = 0.79673 n = 305
What is going on here? Might be related: Why do gam predictions not match gam smooth?

method=REML, as the default GCV is not recommended. – Shawn Hemelstrand Mar 13 '23 at 10:44