I have a question regarding the extracting p-values from the cumulative incidence curves that considered competing risks.
I used cmprsk packages and cuminc function to draw the curves, and
below is the code that I used.
library(cmprsk)
CI.overall <- cuminc(ftime = dt$futime,
fstatus = dt$competingrisks)
CI.overall
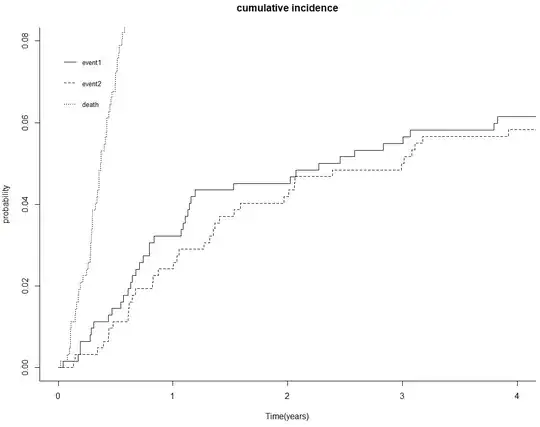
plot(CI.overall, curvlab = c("event1", "event2"),
main= "cumulative incidence", xlab = "Time(years)",
ylab = "probability", xlim=c(0,4), ylim=c(0,0.08),
cex=0.8, lty=c(1,2,0))
variables explanation: competingrisks consists of 0:alive,1:event1,2:event2,3:death (1,2,3 is considered competing risks each other)
The results showed a graph with 3 curves, one curve is for the outcome 1(event1), other curve is for the outcome 2(event2), and the last one is for the death.
and my question is, are there any methods to compare between event1 curve and event2 curve2 and determine a p-value for their being statistically different?
I already know how to compare between 2 groups,for example like sex group, and extract pvalue using code.
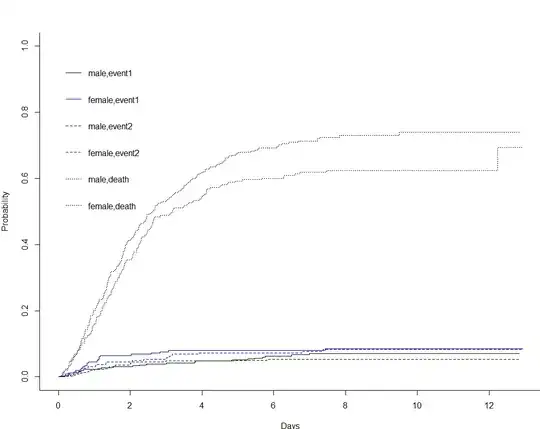
CI.sex <- cuminc(ftime = dt$futime, fstatus=dt$competingrisks,
group=dt$sex)
plot(CI.sex, lty = c(1, 1, 2, 2, 3, 3), col = c("black",
"blue", "black", "blue", "black", "blue"),
curvlab = c("male, event1", "female, event1", "male, event2",
"female,event2", "male,death", "female,death"),
xlab = "Days")
CI.sex$Tests
So,once again, I want to compare between event1 curve and event2 curve2 and extract p-value, and I think it is different from comparing between the 2 groups. In other words, I want to determine whether the cumulative incidence function for event1 differs from that for event2, and just the simple competing-risks model without covariates.
event1differs from that forevent2? Are you modeling any covariate effects, or just the simple competing-risks model without covariates shown in the question? Please provide that information by editing the question, as comments are easy to overlook and can be deleted. – EdM Feb 03 '23 at 17:00