Below you'll find the data and the corresponding R code used to perform the bootstrap hypothesis test to compare the ratio of the means of two samples and additionally, to estimate the 95% confidence interval of such ratios for each of the sample, again via bootstrapping.
X1 <- c(9947.2, 3978.9, 37799, 755.99, 6883.5, 61673, 79577, 15915, 49736, 41800, 31800)
Y1 <- c(5172500, 3163200, 6366200, 915140, 3023900, 1909900, 4894000, 4854200, 3561100, 5829100, 3959000, 2407200, 3779900, 1651200, 3779900)
X2 <- c(216850, 4854.2, 5968.3)
Y2 <- c(65651, 63662, 39789)
D <- mean(X1)/mean(Y1) - mean(X2)/mean(Y2)
boot_D <- rep(0, 10000)
for (i in seq_along(boot_D)) {
boot_D[i] <- mean(sample(X1, replace=TRUE)) / mean(sample(Y1, replace=TRUE)) -
mean(sample(X2, replace=TRUE)) / mean(sample(Y2, replace=TRUE))
}
observed_D <- mean(X1)/mean(Y1) - mean(X2)/mean(Y2)
boot_D.underH0 <- boot_D - mean(boot_D)
mean(abs(boot_D.underH0) > abs(observed_D))
The calculated p-value is:
[1] 0.0928
And the corresponding confidence intervals:
boot_CL1 <- rep(0, 10000)
boot_CL2 <- rep(0, 10000)
for (i in seq_along(boot_CL1)) {
boot_CL1[i] <- mean(sample(X1, replace=TRUE)) / mean(sample(Y1, replace=TRUE))
boot_CL2[i] <- mean(sample(X2, replace=TRUE)) / mean(sample(Y2, replace=TRUE))
}
quantile(boot_CL1, probs = c(.025, .975))
quantile(boot_CL2, probs = c(.025, .975))
2.5% 97.5%
0.004434466 0.013348657
2.5% 97.5%
0.08040818 3.80236249
The confidence intervals do not overlap each other, which makes me wonder why there is a discrepancy between the calculated p-value and the intervals. Shouldn't the p-value in such case be lower than 0.05? If yes, is there an error somewhere in the implementation of either of the procedures?
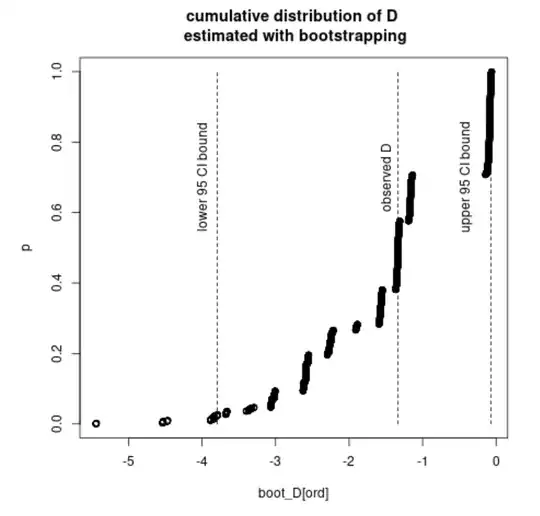
E(X1) / E(Y1), the bootstrap sample ofE(X2) / E(Y2)and finally the bootstrap sample of the difference in ratios. – dipetkov Jan 31 '23 at 18:57