I am running a quasi-poisson regression predicting a count outcome from 4 variables of interest.
I understand that these estimates need to be exponentiated to correctly interpret the results. I used the tab_model() command in sjPlot to gather these and noted that the p-value and associated 95% CI (displayed below) are different from the p-value provided in the glm() output (above).
Could someone please provide any clarification on the discrepancy between these two? Is it typical practice to report the IRR and 95% CI (and possibly whatever this associated p-value is in the tab_model() command)?

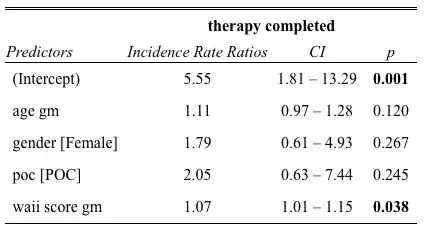
glm()output is derived using Wald tests (https://stats.stackexchange.com/questions/237073/how-does-r-calculate-the-p-value-for-this-binomial-regression). I read that Wald tests may be "valid" for "large samples" but less so in smaller ones, which is arguably not the case in mine (N = 21). Is it reasonable then to report the exponentiated coefficients/CIs and the associated p-values generated viatab_model()? – stilesb Jan 16 '23 at 16:31