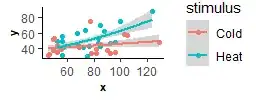
I want to check if there is a significant relationship between x and y, and then check if this relationship is affected by different stimuli (2 levels), colonies (2 levels), and lightings situation (2 levels).
Which statistical test should I apply?
bellow is my sample dataset:
Colony means that individuals are coming from different mothers.
The hypothesis is that under different lighting conditions (whether light or dark), the result will not change.
In this example x is the number of successes during training _ all subjects (ID) have got a specific amount of time (like 10 hours) to do a task, one individual could do it 100 times during the given time, and the other could do only 60 times for example. For y, which is the number of successes during the memory test, all subjects (ID) got a specific amount of time (like 5 hours) to do the task, again one individual did the task 50 times during the given time while another did it only 20 times. So training (x) was done for 10 hours, but the memory (y) was tested for 5 hours, but the timing was the same for all individuals (ID), the number of successes could be as much as an individual is willing to do the task.
df <- structure(list(ID=c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40),
colony=c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2),
stimulus=c("Heat","Heat","Heat","Heat","Heat","Heat","Heat","Heat","Heat","Heat", "Cold","Cold","Cold","Cold","Cold","Cold","Cold","Cold","Cold","Cold","Heat","Heat","Heat","Heat","Heat","Heat","Heat","Heat","Heat","Heat", "Cold","Cold","Cold","Cold","Cold","Cold","Cold","Cold","Cold","Cold"),
lighting=c("light","light","light","light","light","dark","dark","dark","dark","dark","light","light","light","light","light","dark","dark","dark","dark","dark","light","light","light","light","light","dark","dark","dark","dark","dark","light","light","light","light","light","dark","dark","dark","dark","dark"),
x=c(124, 73, 83, 85, 67, 100, 56, 88, 76, 68, 105, 92, 85, 103, 129, 90, 82, 82, 78, 96, 83, 92, 57, 52, 56, 75, 96, 85, 72, 56, 50, 69, 45, 49, 52, 46, 49, 52, 55, 77),
y=c(89, 63, 46, 70, 60, 75, 30, 62, 42, 44, 57, 33, 45, 58, 47, 37, 33, 38, 33, 35, 53, 46, 41, 49, 55, 29, 47, 43, 41, 49, 53, 41, 31, 37, 35, 32, 51, 44, 38, 75)),
class = "data.frame", row.names = c(NA,-40L))
And the main question is whether the number of successes during memory has any relationship to the number of successes during the training (so does having more success during training leads to more success during memory?)