I have some counting data which lists numbers of some incidence in 10 minute intervals.
obs=[1125,1117,1056,...1076] observations in some 112 time intervals.
The data is supposedly Poisson distributed - expecting to see around 1000 incidences in any 10 minutes - but when I try to perform a goodness-of-fit test, I get a p-value of 0.0 --- Now sometimes you simply have to reject your null hypothesis, but I can't help but shake the feeling that I'm doing something wrong, as it's been a while since I had any training in hypothesis testing. The data itself is shown below (with an MLE Poisson pmf plotted on top).
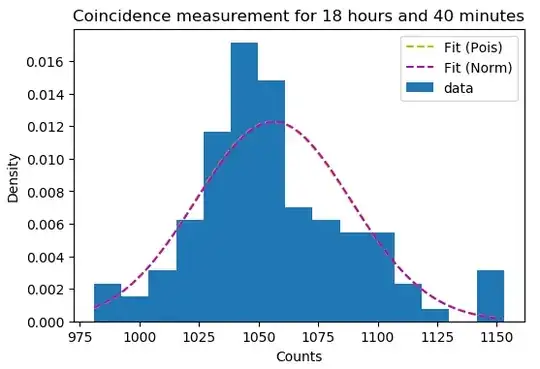
The rate parameter $\lambda$ is estimated with an MLE $\lambda=\overline{n}$, that is; it's just the mean of observations.
I'm using Python and scipy.stats to perform the GoF-test;
from scipy.stats import poisson
from scipy.stats import chisquare
from scipy.stats import chi2
MLE = np.mean(obs)
#H0: The data is Poisson distributed with rate lambda=MLE
#H1: The data is not Poisson distribtued
#under the null hypothesis, the expected values for x observations
with x = 981, 982,..., 1153 (min and max of observations)
Probs = poisson.pmf(obs,MLE)
Ex = Probs * sum(obs)
chisq = sum(((obs-Ex)**2)/Ex) #this tallies up to 253978.198
categories params constant
df=(len(np.unique(coinc))) - 1 - 1
#Chi-squared statistic
chi2.pdf(chisq,df) #this is 0.0
#"automatic version"
chisquare(obs,Ex,df) #returns p-value 0.0
I feel as though I'm messing up by not dividing the data into "categories" in some fashion - as some of the intervals actually do have the same number of counts, for instance the value 1054 occurs three times in the list. Arranging the data into a histogram, however, leaves me a little uncertain how to calculate the expected values (under the null hypothesis).
Edit: Here's the actual data, for testing:
obs = [1125, 1117, 1056, 1069, 1060, 1009, 1065, 1031, 1082, 1034, 985,
1022, 1020, 1108, 1084, 1049, 1032, 1064, 1036, 1034, 1046, 1086,
1098, 1054, 1032, 1101, 1044, 1035, 1018, 1107, 1039, 1038, 1045,
1063, 989, 1038, 1038, 1048, 1040, 1050, 1046, 1073, 1025, 1094,
1007, 1090, 1100, 1051, 1086, 1051, 1106, 1069, 1044, 1003, 1075,
1061, 1094, 1052, 981, 1022, 1042, 1057, 1028, 1023, 1046, 1009,
1097, 1081, 1147, 1045, 1043, 1052, 1065, 1068, 1153, 1056, 1145,
1073, 1042, 1081, 1046, 1042, 1048, 1114, 1102, 1092, 1006, 1056,
1039, 1036, 1039, 1041, 1027, 1042, 1057, 1052, 1058, 1071, 1029,
994, 1025, 1051, 1095, 1072, 1054, 1054, 1029, 1026, 1061, 1153,
1046, 1076]
EDIT: Given the comments, I've tried to redo this with histogram'ing instead. However, I run into a problem with the expectation value for each histogram bin (incidentally, I'm not certain I did it right. Not sure if I should take this question to stackexchange by now...), as some of them are always very low (<1). Generally $\Chi^2$ fits won't work with expectation values below 5 or so; so should I merge the bins before trying to calculate chisq?
# We first arrange data into a histogram
hist,bins,patches=plt.hist(obs,bins=10)
#We need to know which elements went into each bin
#np.digitize returns an array of indices corresponding to which bin from
bins that each value in obs belongs to.
E.g. the first entry is 5; meaning that first entry in obs belongs to
bin number 5
bin_indices=np.digitize(obs,bins)
#under the null hypothesis, the expected values for x observations
Ex=[]
for i in range(len(bins)):
j=np.where(bin_indices==i)[0]
#returns the indices of obs.data which goes in bin number i
data_in_bin=coinc[j]
#returns the coincidence data in that bin, e.g. obs[0] is in bin 5.
if data_in_bin.size >0:
Ex.append(poisson.pmf(data_in_bin,MLE).sum() * sum(hist))
chisq = sum(((hist-Ex)**2)/Ex) #this tallies up to 128.60
categories params constant
df=(len(bins)-1) - 1 - 1
#Chi-squared statistic
p=chi2.pdf(chisq,df) #this is 1.21 e-55
print(p)
#"automatic version"
chisquare(hist,Ex,df) #returns p-value 2.61 e-62
At least some progress was made though. We've gone from $p=0.0$ to $p=1.22\times10^{-55}$.



stats.kstest(obs, 'poisson', [MLE]). Note with large samples these basically always reject, even if the differences with the hypothetical distributions are pretty much trivial for whatever you are doing with the data. – Andy W Dec 08 '21 at 13:28hist,bins,patches=plt.hist(coinc,density=False)yields a list of counts in some intervals, how would one calculate the expected number of counts per bin? Something likeEx = poisson.pmf(center of bin) * sum(hist)where sum(hist) would be 112 (as there are 112 entries inobs) – Woodenplank Dec 08 '21 at 13:57poisson.pmf([1000,1001,1002],1001).sum() * sum(hist). where the list to the pmf function are the integers that the bin includes. – Andy W Dec 08 '21 at 14:55A related question is https://stats.stackexchange.com/questions/12262/
– Sextus Empiricus Jun 01 '23 at 06:24