Simpson's paradox is well known as a situation where the correlation between 2 variables in groups (ie within-group slope) is of opposite sign to the overall correlationed between the 2 variables, ignoring the subgroups (between-group slope)
I have seen several posts where this is illustrated with a simulation. This seems to be a good one: Can adding a random intercept change the fixed effect estimates in a regression model?
Following the code in the above answer:
library(tidyverse)
library(lme4)
set.seed(1234)
n_subj = 5
n_trials = 20
subj_intercepts = rnorm(n_subj, 0, 1)
subj_slopes = rep(-.5, n_subj)
subj_mx = subj_intercepts*2
Simulate data
data = data.frame(subject = rep(1:n_subj, each=n_trials),
intercept = rep(subj_intercepts, each=n_trials),
slope = rep(subj_slopes, each=n_trials),
mx = rep(subj_mx, each=n_trials)) %>%
mutate(
x = rnorm(n(), mx, 1),
y = intercept + (x-mx)*slope + rnorm(n(), 0, 1))
#subject_means = data %>%
group_by(subject) %>%
summarise_if(is.numeric, mean)
subject_means %>% select(intercept, slope, x, y) %>% plot()
Plot
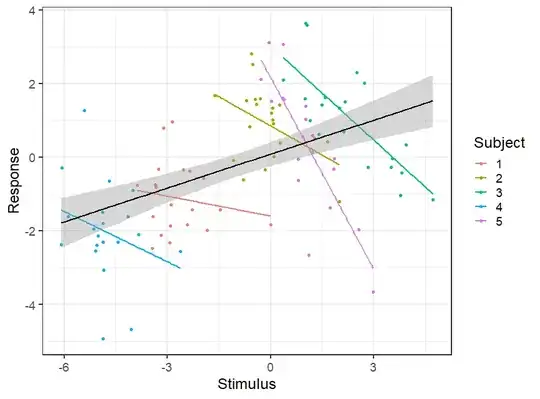
ggplot(data, aes(x, y, color=factor(subject))) +
geom_point() +
stat_smooth(method='lm', se=F) +
stat_smooth(group=1, method='lm', color='black') +
labs(x='Stimulus', y='Response', color='Subject') +
theme_bw(base_size = 18)
The scenario seems quite obvious form the plot. The overall (between-subject) correlation is positive, bue the within-subject correlations are negative. To illustrate this we un an overall regression (lm()) and a regression with random effects (random intercepts for Subject using lmer()):
lm(y ~ x, data = data) %>% summary() %>% coef()
lmer(y ~ x + (1|subject), data = data) %>% summary() %>% coef()
Giving estimates of 0.24 for the between slope and -0.39 for the within slopes. This is good but I thought it would be better if we can see the within and between slopes in the same model. Also the slopes clearly differ quite a lot between subjects, so I thought we could fit the model with random slopes for x:
lmer(y ~ x + (x|subject), data = data) %>% summary() %>% coef()
However this gives a singular fit - correlation between random slopes and intercepts of -1 which does not make sense, so I tried it without the correlation:
lmer(y ~ x + (x||subject), data = data) %>% summary() %>% coef()
but again this is a singular fit because the variance of the random slopes is zero - which also does not make sense because it is clearly quite variable (from the plot).
Advice in this and this post says that we should simplify the random structure. However, that just means goin back to the model with random intercepts only.
So how can we investigate this further and find the within and between subject slopes from the same model ?

Yis simulated with residual variance. – Robert Long Jul 27 '20 at 10:53