The your problem is related to a research field in time series analysis for which there are number of sophisticated methods. But a more abstract problem formulation can use space or any other ordinal variable instead of time.
All the approaches depend on what kind of data you have - the more you know how your data is generated and the context of it the more suitable methods you can find.
Lets break it down in static and dynamic part.
First we are gonna generate some data (i'm using dplyr and ggplot2 packages - but you can handle data with other means):
mydata <- dplyr::bind_rows(
data.frame(time = sample(1:100, size = datasize, replace = T),
value = rnorm(datasize, mean = 100, sd = 10),
group = 1),
data.frame(time = sample(100:200, size = datasize, replace = T),
value = rnorm(datasize, mean = 120, sd = 5),
group = 2),
data.frame(time = sample(200:300, size = datasize, replace = T),
value = rnorm(datasize, mean = 100, sd = 8),
group = 3))
ggplot(mydata, aes(x = time, y = value)) +
geom_point() +
ylim(c(0, 180))
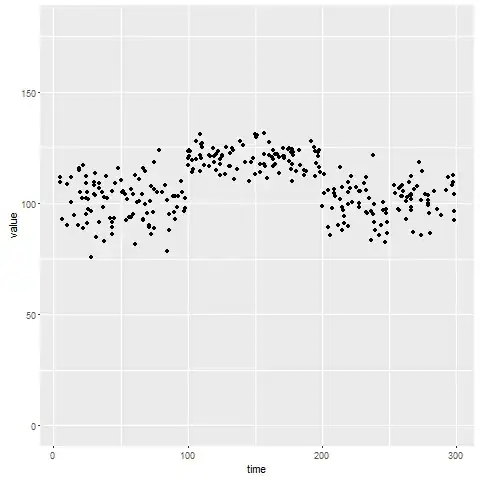
Static approach (and unsupervised)
So you "see" clusters of points that are slightly shifted. That is actually independent of "time" - just consider time and value "static" random variables. What you can do is simple clustering. In order to also get some parameter estimates we can consider the data value as a mixture of Gaussians and use the EM Algorithm (package mixtools required):
mixn <- mixtools::normalmixEM(mydata$value, k = 3, maxit = 10^4)
mixn[c("mu", "sigma")]
$mu
[1] 100.8723 104.5275 120.0161
$sigma
[1] 9.5199389 0.9243747 5.1829086
The means are more or less captures accurately, but the sigmas not so much. Anyhow, your mixture would look like this:

You would then, for every point, compute its likelihood belonging to either distribution - and choose a class with the maximal likelihood.
Furthermore, we can compute more precise clusters using value and time variable togther - again from a static point of view.
mixmvn <- mixtools::mvnormalmixEM(as.matrix(mydata[c("time", "value")]), k = 3)

Although the result seems nice, you need to deal with covariance matrices for interpretation.
Dynamic approach (and supervised)
What you may looking for are Hidden Markov Models. In their basic version they model discrete states with discrete observations. They can be extended to continous observations. (Btw: continous state and constinous obeservations are related to the topic of Kalman-filters).
To estimate the parameters we can use package RHmm which atm can only be installed from achive RHmm archive by R CMD INSTALL R_Hmm_2.0.3.tar.gz.
# we need to order the values by time
orderedValues <- (mydata %>% dplyr::arrange(time))$value
hmm <- RHmm::HMMFit(obs = orderedValues, nStates = 3, dis = "NORMAL", list(iter = 1000, nInit = 100), asymptCov = F)
The results are very nice:
> hmm$HMM$distribution
Model:
3 states HMM with univariate gaussian distributions
Distribution parameters:
mean var
State 1 120.28424 24.27617
State 2 97.56794 72.53311
State 3 106.79554 31.86263
> sqrt(hmm$HMM$distribution$var) # sigma
[1] 4.927085 8.516637 5.644699
The means as well as sigmas are estimated more or less correctly. We can use predict for new samples in order to assign a state if we want.
All these methods has a distadvantage, however. They all need a fist estimate of number states.
If you are only interested in "change" in dynamic (and have a lot of data), a Hebb's rule may be your friend. Afaik Hebb's rule get accustomed to your data and you will see a spike if the sequential input changes "dramatically".
Any thoughts ?



