I have an image data matrix $X \in \Re^{N \ \text{x}\ p}$ where $N=50000$ is the number of image examples, and $p=3072$ is the number of image pixels: $p = 3072 = 32 \times 32 \times 3$, because each image is a 3-channel $32 \times 32$ image. Furthermore, each of the 50000 images belongs to 1 of 10 possible classes. That is, there are 5000 images of class 'car', 5000 images, of class 'bird', etc... and there are 10 classes total. This is a part of the CIFAR-10 dataset.
The ultimate goal here is to perform classification on this data set. To this end, the professor mentioned to try PCA on this, and then placing those features into a classifier. As my classifier, I am using a fully connected neural network with one hidden layer and a softmax output.
My problem is that I believe I have done PCA in the correct way, but I think my way might possibly be mis-applied.
This is what I have done:
In order to compute the PCA of my data, this is what I did so far:
First, I compute the mean image $\mu \in \Re^{1\text{x} p}$. Let $x_n$ be the $n$'th row of $X$. Then,
$$ \mu = \frac{1}{N} \sum_{n=1}^{N} x_n $$
Compute the covariance matrix $C \in \Re^{p \text{x} p}$ of my image data:
$$ C = \frac{1}{N-1}(X - \mu)^{T}(X - \mu) $$
Perform an eigen-vector decomposition of $C$, yielding $U$, $S$, and $V$, where the matrix $U$ encodes the principal directions (eigenvectors) as columns. (Also, assume that eigen-values are already sorted in decreasing order). Thus:
$$ [U, S, V] = \text{eig}(C) $$
Finally, perform PCA: i.e, compute a new data matrix $P \in \Re^{N \text{x} k}$, where $k$ is the number of principal components we wish to have. Let $U_k \in \Re^{p \text{x} k}$ - that is, a matrix with only the first $k$ columns. Thus:
$$ P = XU_k $$
The Question:
I think my method of performing PCA on this data is mis-applied, because the way I have done it, I basically end up decorrelating my pixels from each other. (Assume I had set $k=p$). That is, the resultant rows of $P$ look more or less like noise. That being the case, my questions are as follows:
- Have I really de-correlated the pixels? That is, have I in fact removed any coupling between pixels that a prospective classifier might have hoped to use?
- If the answer to the above is true, then why would we ever do PCA this way?
- Finally, related to the last point, how would we do dimensionality reduction via PCA on images, if in fact, the method I have used it wrong?
EDIT:
After further studying and plenty of feedback, I have refined my question to: If one were to use PCA as a pre-processing step for image classification, is it better:
- Perform classification on the k principal components of the images? (Matrix $X_{new} = X U_k$ in the above, so now each image is of length $k$ instead of the original $p$)
- OR is it better to perform classification on the reconstructed images from k-eigenvectors, (which will then be $X_{new} = X U_k U_k^T$, so even though each each image is STILL the original $p$ in length, it was in fact reconstructed from $k$ eigenvectors).
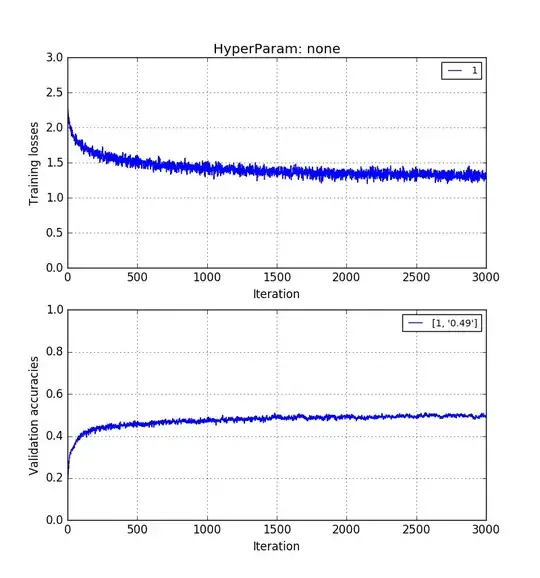
Empirically, I have found that validation accuracy without PCA > validation accuracy with PCA reconstruction > validation accuracy with PCA PCs.
The images below show that in the same order. 0.5 > 0.41 > 0.31 validation accuracies.
Training on raw pixels images of length $p$:
Training on images of length $p$ but reconstructed with k=20 eigenvectors:
And finally, training on the $k=20 principal components themselves*:
All this has been very illuminating. As I have found out, PCA makes no guarantees that the principal components make demarcation between different classes easier. This is because the principal axes computed are axes that merely try to maximize the energy of projection across all images, agnostic to image class. In contrast, actual images - whether faithfully reconstructed or not, still maintain some aspect of spatial differences that can go - or should go - towards making classification possible.


eig()are not necessarily sorted. Your notation of[U,S,V]would normally be used forsvd(), which does return sorted results. (In either case, you should have V=U, due to symmetric input; the full SVD could be used on X, but not on X'*X). I ask because if the output is not sorted, any patterns could be hidden. – GeoMatt22 Sep 08 '16 at 02:13svd()would be needed for $X$, as it is not a symmetric matrix. Note that if we assume for simplicity that $X$ is centered (0-mean), the covariance is $C=X^TX$. Then given[U,S,V]=svd(X), i.e. $X=USV^T$, the covariance decomposition will be $C=V(S^2/N)V^T$. For grayscale images, commonly the $k$ roughly corresponds to "frequency" (more oscillatory basis vectors), and the energy decays with $k$. Not sure if this holds for you? The RGB stack could change things also. – GeoMatt22 Sep 08 '16 at 02:30X*UandX_newcontain exactly the same information up to a linear transform, which can be taken care of in the first layer mapping. You should be getting identical performance. (In any case, I think you should update your question with these important details.) – amoeba Sep 12 '16 at 22:15