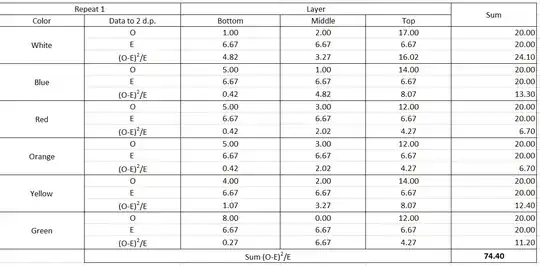
You shouldn't be using a 'one-way' or 'goodness-of-fit' chi-squared test here six times over. You should be using a chi-squared test of independence on a two-way contingency table. In addition, as @DJohnson notes below, you need to use the actual counts observed, not average counts (I'm not sure I understand how you say you got $6.67$ flies in the bottom layer, for example.) That is, you need to set up a contingency table like this:
Layer
Color bottom middle top sum
red 7 3 10 20
green # # # 20
blue # # # 20
orange # # # 20
purple # # # 20
yellow # # # 20
Then run your chi-squared test. The degrees of freedom for chi-squared test is $(r-1)(c-1)$ (i.e., the number of rows minus 1 times the number of columns minus 1). In your case that would be: $5\times 2 = 10$.
Update: If you have three repeated versions of this experiment, you have (in some sense) three two-way contingency tables, or (more correctly) a three-way contingency table. You want to test if there is a difference amongst the rows with the iterations taken into account. The general way to analyze mult-way contingency tables is to use the log linear model (which is actually a dressed-up Poisson GLiM). I describe this in more detail here: $\chi^2$ of multidimensional data. Below, I create two fake datasets using R, one I call ".n" (for 'null', because there isn't a relationship between the color and the layer), and the other I call ".a" (for 'alternative', because the relationship you are interested in does exist).
dft = expand.grid(layer=c("bottom","middle","top"),
color=c("blue", "green", "orange", "red", "white", "yellow"),
Repeat=1:3)
dft = dft[,3:1]
dft.n = data.frame(dft, count=c(rep(c( 3,6,11), times=6),
rep(c( 6,7, 7), times=6),
rep(c(11,6, 3), times=6)))
dft.a = data.frame(dft,
count=c(c(3,6,11), c(11,6, 3), c(11,6, 3), c(3,6,11), c(3,6,11), c(11,6, 3),
c(3,6,11), c(11,6, 3), c(11,6, 3), c(3,6,11), c(3,6,11), c(11,6, 3),
c(3,6,11), c(11,6, 3), c(11,6, 3), c(3,6,11), c(3,6,11), c(11,6, 3) ))
tab.n = xtabs(count~color+layer+Repeat, dft.n)
# , , Repeat = 1
# layer
# color bottom middle top
# blue 3 6 11
# green 3 6 11
# orange 3 6 11
# red 3 6 11
# white 3 6 11
# yellow 3 6 11
#
# , , Repeat = 2
# layer
# color bottom middle top
# blue 6 7 7
# green 6 7 7
# orange 6 7 7
# red 6 7 7
# white 6 7 7
# yellow 6 7 7
#
# , , Repeat = 3
# layer
# color bottom middle top
# blue 11 6 3
# green 11 6 3
# orange 11 6 3
# red 11 6 3
# white 11 6 3
# yellow 11 6 3
tab.a = xtabs(count~color+layer+Repeat, dft.a)
# , , Repeat = 1
# layer
# color bottom middle top
# blue 3 6 11
# green 11 6 3
# orange 11 6 3
# red 3 6 11
# white 3 6 11
# yellow 11 6 3
#
# , , Repeat = 2
# layer
# color bottom middle top
# blue 3 6 11
# green 11 6 3
# orange 11 6 3
# red 3 6 11
# white 3 6 11
# yellow 11 6 3
#
# , , Repeat = 3
# layer
# color bottom middle top
# blue 3 6 11
# green 11 6 3
# orange 11 6 3
# red 3 6 11
# white 3 6 11
# yellow 11 6 3
I run a quickie log-linear analysis on both. The models are listed from 0, which is the 'saturated' model, through 2, which has dropped terms. Note that in R it is typical to list models in order from smallest to largest, but the result of the anova() call refers to the nested model as "Model 1", which makes the names not correspond well; try not to be thrown off by this. For the null dataset, Model 2 differs from Model 1 (i.e., m.1.n differs from m.2.n), meaning that the layers are not independent of the Repeats. On the other hand, Model 3 does not differ from Model 2 (i.e., m.0.n differs from m.1.n), meaning that the layer*Repeat pattern does not differ by color. In addition, Model 3 does not differ from the Saturated model (because it is the saturated model).
library(MASS)
m.0.n = loglm(~color*layer*Repeat, tab.n)
m.1.n = loglm(~color+layer*Repeat, tab.n)
m.2.n = loglm(~color+layer+Repeat, tab.n)
anova(m.2.n, m.1.n, m.0.n)
# LR tests for hierarchical log-linear models
#
# Model 1:
# ~color + layer + Repeat
# Model 2:
# ~color + layer * Repeat
# Model 3:
# ~color * layer * Repeat
#
# Deviance df Delta(Dev) Delta(df) P(> Delta(Dev)
# Model 1 59.55075 44
# Model 2 0.00000 40 59.55075 4 0
# Model 3 0.00000 0 0.00000 40 1
# Saturated 0.00000 0 0.00000 0 1
m.0.a = loglm(~color*layer*Repeat, tab.a)
m.1.a = loglm(~color+layer*Repeat, tab.a)
m.2.a = loglm(~color+layer+Repeat, tab.a)
anova(m.2.a, m.1.a, m.0.a)
# LR tests for hierarchical log-linear models
#
# Model 1:
# ~color + layer + Repeat
# Model 2:
# ~color + layer * Repeat
# Model 3:
# ~color * layer * Repeat
#
# Deviance df Delta(Dev) Delta(df) P(> Delta(Dev)
# Model 1 87.47794 44
# Model 2 87.47794 40 0.00000 4 1e+00
# Model 3 0.00000 0 87.47794 40 2e-05
# Saturated 0.00000 0 0.00000 0 1e+00
For the alternative dataset, Model 2 does not differ from Model 1 (i.e., m.1.a differs from m.2.a), meaning that the layers are independent of the Repeats. On the other hand, Model 3 does differ from Model 2 (i.e., m.0.a differs from m.1.a), meaning that the layer*Repeat pattern does differ by color. (And again, Model 3 is the Saturated model.)