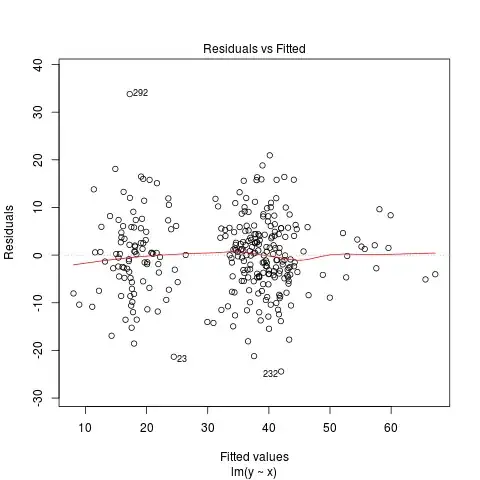
I am checking that I have met the assumptions for multiple regression using the built in diagnostics within R. I think that from my online research, the DV violates the assumption of homoscedasticity (please see the residuals vs fitted plot below).

I tried log transforming the DV (log10) but this didn't seem to improve the residuals vs fitted plot. There are 2 dummy coded variables within my model and 1 continuous variable. The model only explains 23% of the variance in selection (DV) therefore, could the lack of homoscedasticity be because variable/s are missing? Any advice on where to go from here would be greatly appreciated.
plot(lm(y~x)). – C.R. Peterson Nov 18 '15 at 04:18fm=lm(y~x);plot(y~fitted(fm)), but you can usually figure out what it will look like from the residual plot -- if the raw residuals are $r$ and the fitted values are $\hat{y}$ then $y$ vs $\hat{y}$ is $r + \hat{y}$ vs $\hat{y}$; so in effect you just skew the raw residual plot up 45 degrees. – Glen_b Nov 18 '15 at 04:29mtcars):fit <- lm(mpg ~ wt, mtcars); Predicted <- predict(fit); plot(mtcars$mpg ~ Predicted, ylab="Observed", main = "Observed v. Fitted"); abline(0,1). Please post it if you get it! – Antoni Parellada Nov 18 '15 at 19:01