I'm using GPML toolbox by C.E.Rasmussen to solve the basic GP regression problem (presented in the book) with noisy observations. That is to say, estimate the underlying function $f$ of a static noisy mapping
$$y = f(\mathbf{x}) + e, \qquad e \sim \mathcal{N}(0, \sigma^2)$$
from a set of training examples $\{ (\mathbf{x}_i, y_i) \}_{i=1}^{n}$. As far as I understand it, I should respect the noisiness of observations by choosing the kernel as the sum
$$ k(\mathbf{x}_i, \mathbf{x}_j) = k_f(\mathbf{x}_i, \mathbf{x}_j) + \sigma^2_{e}\delta_{ij}$$
where the final term in the sum is the kernel of the white noise (that is, noise of the observations).
When using GPML toolbox, for those who are familiar, you have to specify a likelihood. In my case I chose Gaussian likelihood, which has one hyperparameter - in the code documentation this correponds to formal parameter $s_n$.
So all together, when I perform optimization, I have one hyperparameter for the noise kernel ($\sigma_e$), one for the likelihood ($s_n$) and $d$ (say) hyperparameters for the $k_f$.
I am confused about the meaning of the hyperparameters $\sigma_e$ and $s_n$. Which one of the hyperparameters ($\sigma_e$ or $s_n$) represents the variance of the noise in the observations?
If the Gaussian likelihood is the measurement model, then $s_n$ should be the variance of the observations $y_i$, but then for what reason do we add the noise kernel (with additional hyperparameter ($\sigma_e$), which I think is redundant, at this point since we already have $s_n$ to do the job)? Perhaps they're one and the same and should be tied together during optimization. I'm confused.
GPML code for exact inference:
[n, D] = size(x);
K = feval(cov{:}, hyp.cov, x); % evaluate covariance matrix
m = feval(mean{:}, hyp.mean, x); % evaluate mean vector
sn2 = exp(2*hyp.lik); % noise variance of likGauss
if sn2<1e-6 % very tiny sn2 can lead to numerical trouble
L = chol(K+sn2*eye(n)); sl = 1; % Cholesky factor of covariance with noise
pL = -solve_chol(L,eye(n)); % L = -inv(K+inv(sW^2))
else
L = chol(K/sn2 + eye(n)); sl = sn2; % Cholesky factor of B
pL = L; % L = chol(eye(n)+sW*sW'.*K)
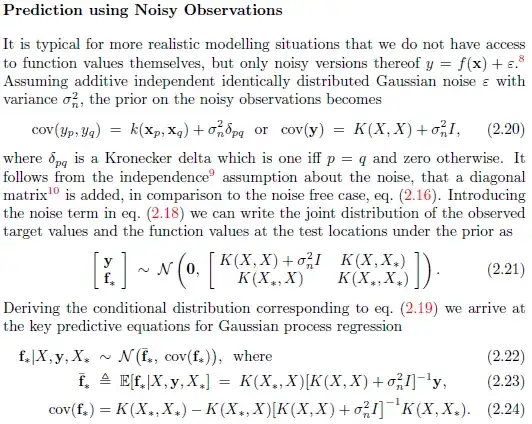
end
alpha = solve_chol(L,y-m)/sl;
sn2 is the likelihood parameter, hyp.cov contains the kernel hyperparameters (including the noise kernel hyperparameter $\sigma_e$)