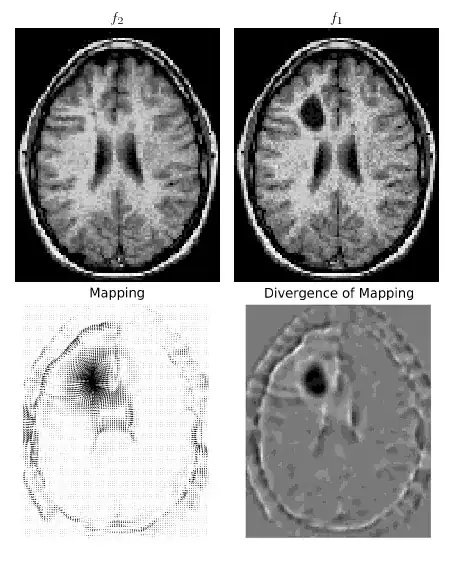
I am implementing the paper "Optimal Mass Transport for Registration and Warping", my goal being to put it online as I just cannot find any eulerian mass transportation code online and this would be interesting at least for the research community in image processing.
The paper can be summarized as follows :
- find an initial map $u$ using 1D histogram matchings along the x and y coordinates
- solve for the fixed point of $u_t = \frac{1}{\mu_0} Du \nabla^\perp\triangle^{-1}div(u^\perp)$ , where $u^\perp$ stands for a 90 degrees counter clockwise rotation, $\triangle^{-1}$ for the solution of the poisson equation with Dirichlet boundary conditions (=0), and $Du$ is the determinant of the Jacobian matrix.
- stability is guaranteed for a timestep $dt<\min|\frac{1}{\mu_0}\nabla^\perp\triangle^{-1}div(u^\perp)|$
For numerical simulations (performed on a regular grid), they indicate using matlab's poicalc for solving the poisson equation, they use centered finite differences for spatial derivatives, except for $Du$ which is computed using an upwind scheme.
Using my code, the energy functional and curl of the mapping are properly decreasing for a couple iterations (from a few tens to a few thousands depending on the time step). But after that, the simulation explodes : the energy increases to reach a NAN in very few iterations. I tried several orders for the differentiations and integrations (a higher order replacement to cumptrapz can be found here), and different interpolation schemes, but I always get the same issue (even on very smooth images, non-zero everywhere etc.).
Anyone would be interested in looking at the code and/or the theoretical problem I am facing ? The code is rather short.
Code with debugging functions
Just the necessary function without test stuffs (< 100 lines)
Please replace gradient2() at the end by gradient(). This was a higher order gradient but doesn't solve things either.
I am only interested in the optimal transport part of the paper for now, not the additional regularization term.
Thanks !