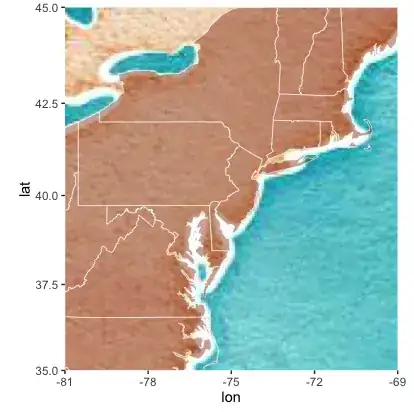
I'm trying to incorporate a shapefile of US states with ggmap and keep getting "tearing" of my polygons. I tried changing group=id to group=group which helped, but didn't seem to resolve the boundary.
library(rgdal)
states <- readOGR(dsn="shapefiles", layer="states")
proj4string(states)
\[1] "+proj=longlat +datum=NAD83 +no_defs +ellps=GRS80 +towgs84=0,0,0"
states <- spTransform(states, CRS("+proj=longlat +datum=WGS84"))
states <- fortify(states)
sstates <- get_map(location = c(-81, 35, -69, 45), zoom = 4, maptype = "watercolor")
sstates <- ggmap(sstates)
sstates <- sstates + geom_polygon(aes(x = long, y = lat, group=group),
data = states, color ="white", fill ="orangered4",
alpha = .4, size = .2)
Original shapefile
https://www.arcgis.com/home/item.html?id=f7f805eb65eb4ab787a0a3e1116ca7e5