I have found a couple of sources1,2 that indicate that a read in a 1D² run is classified by MinKNOW as "pass" and put into the fastq_pass folder if both of the following conditions are met:
- Both strands were read
- The average Phred score of all bases in the read was >9
(Thus a read in a 1D² run goes into the fastq_fail folder if either its Phred is too low or if the second strand didn't follow the first through the pore.)
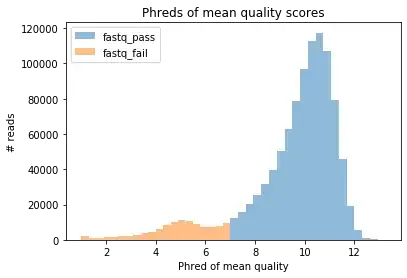
However, I have been unable to track down any source indicating how the pass/fail classification works for 1D runs (where only one strand is supposed to be read per read). One might reasonably guess that it's based solely upon whether the mean Phred score is above 9, but inspection of the data from a 1D run reveals that this is not the case. While all "pass" reads seem to have a mean Phred score of at least 9 (the lowest I see is 9.45), some fail reads have scores above 9 too (with the highest one being 12.9):
Thus it looks to me like this is not the condition used to classify 1D reads. But then what is?