Does anyone know where and how could I download a list of all human genes and the length of the coding sequence for each gene? Is it possible to do this on the NCBI site, ensembl?
-
2Which coding sequence? I mean, do you just want whichever has been designated the 'canonical' transcript or do you want all possible isoforms? – terdon Apr 30 '19 at 14:54
-
Hi terdon, thanks for the quick reply! Yes exactly, the canonical transcript is good enough! – solimanelefant Apr 30 '19 at 14:56
-
1Michael G. suggests to take a look at relevant front-end, NCBI's eFetch. Which is supposedly perfect for what you need. – Kamil S Jaron May 01 '19 at 07:53
3 Answers
While I haven't found a way to limit the results to the canonical transcript only, you can get a list of genes, transcripts and their CDS lengths using Ensemble's BioMart. I have already set it up for you, you can see the results, and modify them, here (click on the "Results" link if you don't see them).
Essentially, you just need to go to BioMart, and
select "Ensembl Genes 96" (the number will change if the version changes) as the database and "uman Genes" as the dataset.
Click on "Filters", and set
Gene typetocodingandTranscript typetoprotein_coding.From "Attributes", select whatever you want to see. The "CDS Length" is under "Structures".
- 10,071
- 5
- 22
- 48
Go to the NCBI homepage and search for 'human genome' (the dropdown menu on the left-hand side of the search box should be 'All Databases'). In the result page, click on the 'Download' button; choose 'RefSeq' as the 'Source database' and 'Feature table' as the 'File type' as shown in the screenshot below:
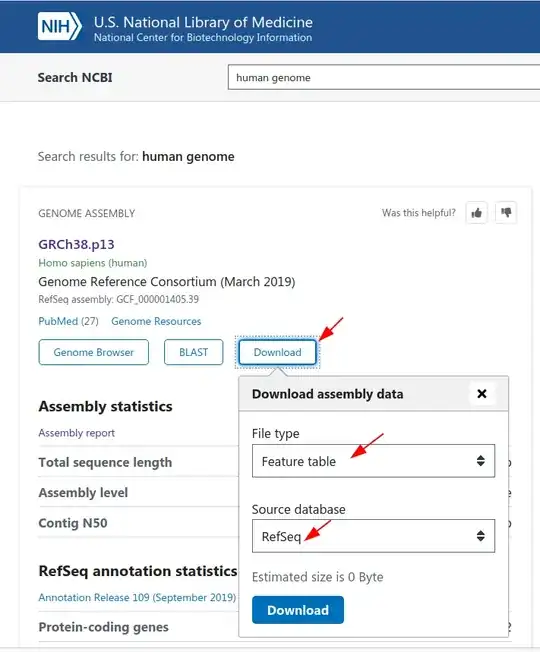
This will download a genome_assemblies.tar file which contains the feature table file, GCF_000001405.39_GRCh38.p13_feature_table.txt.gz. This file has all of the annotated features in a tab-delimited format with the following fields:
# feature [ 1]: gene
class [ 2]: transcribed_pseudogene
assembly [ 3]: GCF_000001405.39
assembly_unit [ 4]: Primary Assembly
seq_type [ 5]: chromosome
chromosome [ 6]: 1
genomic_accession [ 7]: NC_000001.11
start [ 8]: 11874
end [ 9]: 14409
strand [ 10]: +
product_accession [ 11]:
non-redundant_refseq [ 12]:
related_accession [ 13]:
name [ 14]: DEAD/H-box helicase 11 like 1
symbol [ 15]: DDX11L1
GeneID [ 16]: 100287102
locus_tag [ 17]:
feature_interval_length [ 18]: 2536
product_length [ 19]:
attributes [ 20]: pseudo
You can parse this file to obtain the CDS lengths of all annotated proteins. As an example, you can get a list of all annotated RefSeq proteins and their lengths as follows:
zcat GCF_000001405.39_GRCh38.p13_feature_table.txt.gz \
| awk 'BEGIN{FS="\t";OFS="\t"}($1~/^#/ || $1=="CDS")' \
| cut -f6-11,13,15,16,18,19
The output of the above command will have the following fields:
chromosome [ 1]: 1
genomic_accession [ 2]: NC_000001.11
start [ 3]: 69091
end [ 4]: 70008
strand [ 5]: +
product_accession [ 6]: NP_001005484.1
related_accession [ 7]: NM_001005484.1
symbol [ 8]: OR4F5
GeneID [ 9]: 79501
feature_interval_length [ 10]: 918
product_length [ 11]: 305
- 1,266
- 6
- 9
Ensembl has an FTP site that allows you to select and download only the coding sequences from many different genomes. https://useast.ensembl.org/info/data/ftp/index.html
To determine the length of those sequences, download the associated gtf or gff3 annotation file. The annotation file is tab delim. The fourth and fifth column represent genomic loci of the annotated region. Subtract the amount in the fourth column from the amount in the fifth column to yield the length of all the annotated features. You can easily do this in R after loading the file in the environment using the Magrittr library. The following code will create a new column called gene_length with the associated gene lengths.
install.packages("magrittr")
# this only needs to be done once
library(magrittr)
# must be run each time the library is neaded
annotation.gtf <- read.table("path/to/annotation.gtf")
annotation.gtf$start <- annotation.gtf[,4]
annotation.gtf$end <- annotation.gtf[,5]
annotation.new-column.gtf <- annotation.gtf %>%
mutate(gene_length=end-start)
- 5,542
- 2
- 25
- 59
- 41
- 4