Currently I'm performing whole analysis (pipeline from *.fastq to *.vcf) of 41 samples (targeted NGS). I rely on GATK best practices, however with some modifications. I decided to use the following tools:
#mapping
bwa mem (with mem - alternate haplotypes)
#preprocessing
RevertSam (Picard)
MergeBamAlignment (Picard)
MarkDuplicates (Picard)
BaseRecalibrator (GATK)
ApplyBQSR (GATK)
AnalyzeCovariates (GATK)
SortSam (Picard)
#call, filter and annotate variants
HaplotypeCaller (GATK)
GenotypeGVCFs (GATK)
snpEff, SnpSift
Nevertheless, after marking the duplicates with MarkDuplicates about 99% of reads have been marked as duplicated, hence the coverage decreased from several hundreds to literally up to five. So I decided to omit this step.
Have you ever encountered a similar problem? What is your opinion about this?
Best Regards, Adam
EDIT
The Agilent Haloplex custom target enrichment kit (HaloPlex Target Enrichment for Illumina Kit) has been used to prepare the libraries, sequencing done using Illumina HiSeq 2500. Targeted NGS of all exons (only) of 90 genes.
Here I provide how the final bam look like. This is the result without duplicate marking, otherwise there're only several reads per exon...
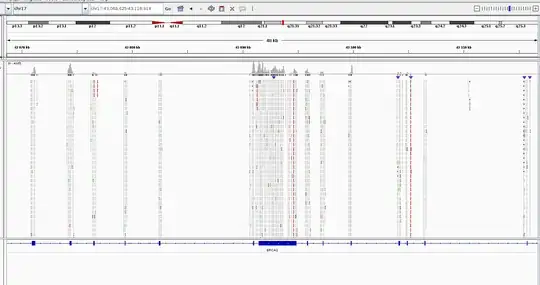
MarkDuplicatesthe coverage was up to 4. It was hard to distinguish between actual alternations and mismatched bases. – Adamm Feb 15 '19 at 05:55