I am analysing singe cell sequence data and I have followed this tutorial, https://satijalab.org/seurat/pbmc3k_tutorial.html to perform QC and various differential analyses using the Seurat package on my data up till now.
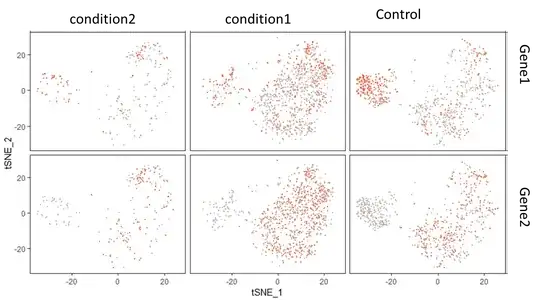
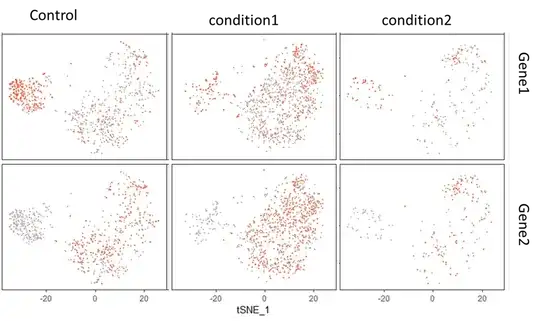
How can I rearrange on the resulting plot, the order in which the groups of my samples appear when I use the FeatureHeatmap() function to visualise the differential gene expression of Gene1 and Gene2 across the cell clusters?
FeatureHeatmap(object = object, features.plot = c("Gene1", "Gene2"),
group.by = "Conditions",
pt.size = 0.25, key.position = "top",
max.exp = 3)
VariableFeaturesis not present in my Seurat package. I get the error:Error in VariableFeatures(object) : could not find function "VariableFeatures"@TimStuart – Charles Jan 18 '19 at 14:26devtools::install_github(repo = 'satijalab/seurat', ref = 'release/3.0'). I don't know how much effort it is for you to repeat analyses with Seurat v3, but you can also just update the Seurat object usingUpdateSeuratObject. See here for more information: https://satijalab.org/seurat/essential_commands.html – TimStuart Jan 18 '19 at 16:30devtools::install_github(repo = 'satijalab/seurat', ref = 'release/3.0'):package ‘curl’ successfully unpacked and MD5 sums checked: Error: (converted from warning) cannot remove prior installation of package ‘curl’.UpdateSeuratObject(object): Object representation is consistent with the most current Seurat version.The
FeatureHeatmap()actually uses the alphabetical order of the conditions which I provided in the metadata slot. Is there a possibility to reorder the factor levels without harming the integrity of the data in the Seurat object? – Charles Jan 20 '19 at 15:11UpdateSeuratObject? – TimStuart Jan 21 '19 at 14:33