I have short (67bp) Hi-C reads from a highly heterozygous organism (~15% between-haplotype SNP divergence) and I have both reference haplotypes.
I wanted to try and compare different haplotyping softwares for Hi-C reads using these reads as benchmarking dataset. When mapping the reads separately to each haplotype, I get good mapping statistics. When I map the reads to a single haplotype with all the heterozygous SNPs masked (into N's), I get very poor mapping rates.
I would like to be able to map the reads when the real haplotypes are unknown (reference is a mix of haplotypes).
I use minimap2 to map the reads with the sr preset. I tried decreasing the mismatch penalty (-B) to 1 and increasing the --score-N value, but this had no effect.
As shown in the attached IGV screenshot, coverage drops to 0 when the SNP density increases.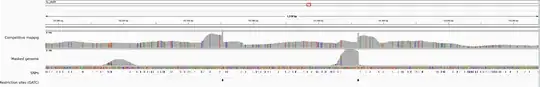 Is it feasible to map reads with such a high heterozygosity on a single (masked) reference ? Should I use another tool ?
Is it feasible to map reads with such a high heterozygosity on a single (masked) reference ? Should I use another tool ?