I have a set of scRNA-seq samples expressing TdTomato, which has high content in microscope. I followed the 10x cellranger pipelines to finsh the work, my procedures are as follows:
- cellranger mkref
- cellranger count
I got the result, which includes:

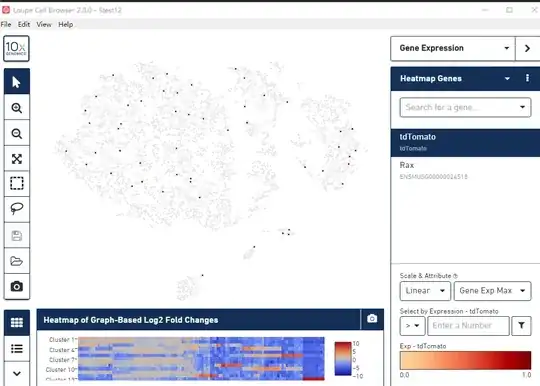
But strangely, the tdtomato expression content in my three sample are all very low! And cells with tdtomato are also very low and disperse!! Take sample 1 as example, you can see the picture:
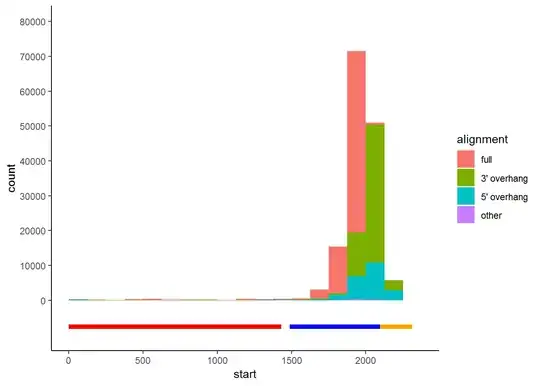
It's not normal, but I can't find the reason. Following the 10x genomics help documentation noticed that by executing the CellRanger filtering steps, maybe my tdtomato reads are filtered too much, but because I'm not very skillful at programming and I'm first time to do RNA-seq (and scRNA-seq), I just don't know how to analysis the bam file as suggested by the official website and find the real reason of the problem:
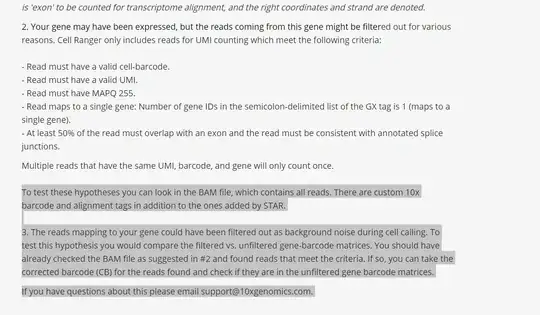
If you can show me the code need to do, I will be very grateful!