I have a set of differentially methylated/expressed/whatever entities with p-values attached (example below).
entity_name p-value magnitude
entity1 0.04459 0.68
entity2 0.02283 0.99
...
entity_n 0.78 0.025
Typically, I apply the p.adjust function in R with the "fdr" (Benjamini-Hochberg) approach to leave me with p-values adjusted to control the FDR.
adjusted <- p.adjust(mydata,"fdr")
However, I am interested in showing a volcano plot with the unadjusted p-values, and two alpha levels: 0.05 and one that corresponds to the correction. What is the best way to get this alpha? Is it appropriate to set the "corrected alpha" to the lowest original p-value that doesn't pass FDR correction?
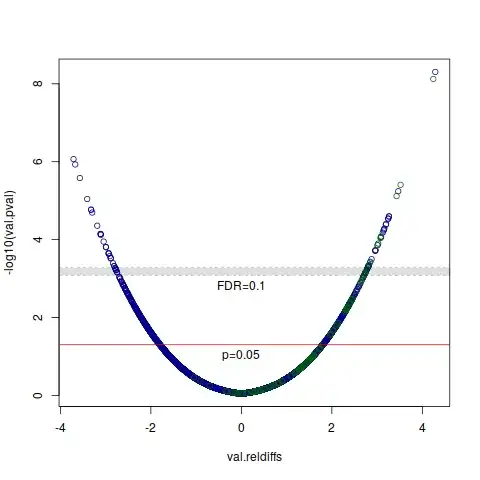
< 0.05, not<= 0.05. – Konrad Rudolph Jun 13 '17 at 10:20