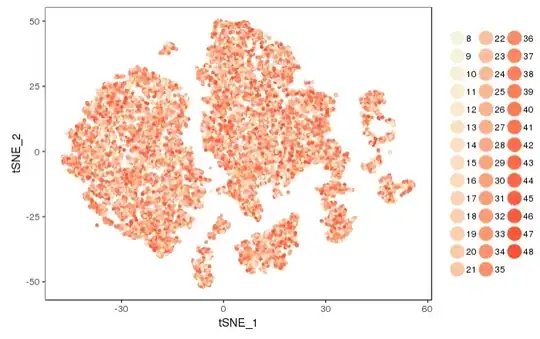
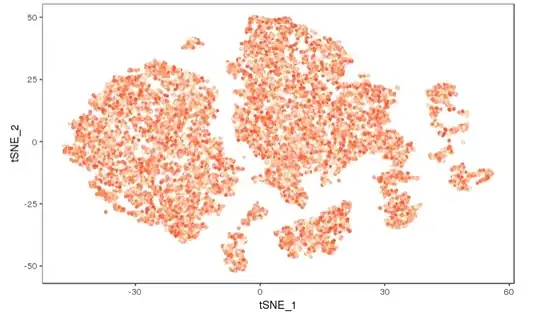
I returned a FeaturePlot from Seurat to ggplot. My plot has a weird range of colours as below
I produced this plot by this code
> head(mat[1:4,1:4])
s1.1 s1.2 s1.3 s1.4
DDB_G0267178 0 0.009263254 0 0.01286397
DDB_G0267180 0 0.000000000 0 0.00000000
DDB_G0267182 0 0.000000000 0 0.03810585
DDB_G0267184 0 0.000000000 0 0.00000000
>
I have converted expression matrix to a binary matrix by 2 as a threshold
mat[mat < 2] <- 0
mat[mat > 2] <- 1
> head(exp[1:4,1:4])
s1.1 s1.2 s1.3 s1.4
DDB_G0267382 0 0 0 1
DDB_G0267438 0 0 0 1
DDB_G0267466 0 0 0 0
DDB_G0267476 0 0 1 0
>
> exp=colSums(exp)
> exp=as.matrix(exp)
> colnames(exp)="value"
> exp=as.data.frame(exp)
> cc <- AddMetaData(object = seurat, metadata = exp)
> cc <- SetAllIdent(object = cc, id = "value")
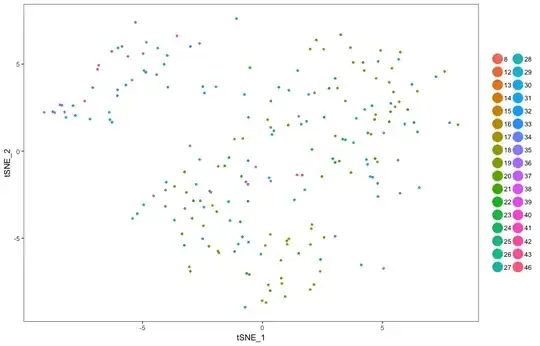
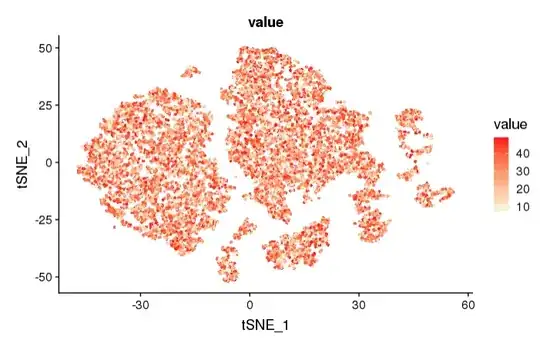
> TSNEPlot(object = cc, do.return= TRUE)
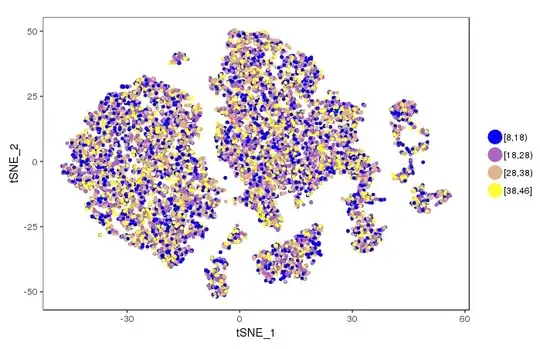
How I can convert this range to a gradient of colours for example in 8-18, 18-28, 28-38, 38-48 range in blue to yellow please? Something like below
Thank you for any help
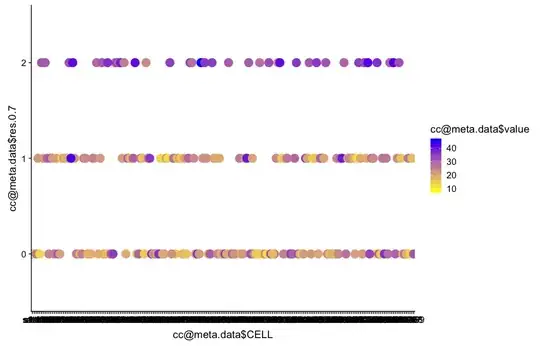
Then by ggplot now I scaled my colours but I don't like my clusters as so and I don't know how to retain my clusters as a featureplot by this new color gradient
> head(cc@meta.data)
nGene nUMI orig.ident res.0.7 CELL STAGE GENO dataset stage.nice celltype value
s1.1 4331 373762 SeuratProject 0 s1.1 H16 WT 1 H16 0 34
s1.2 5603 1074639 SeuratProject 0 s1.2 H16 WT 1 H16 0 26
s1.3 2064 49544 SeuratProject 0 s1.3 H16 WT 1 H16 0 27
s1.4 4680 772399 SeuratProject 1 s1.4 H16 WT 1 H16 1 29
s1.5 3876 272356 SeuratProject 1 s1.5 H16 WT 1 H16 1 21
s1.6 2557 122314 SeuratProject 0 s1.6 H16 WT 1 H16 0 31
> ggplot(as.data.frame(cc@meta.data), aes(x = cc@meta.data$CELL, y = cc@meta.data$res.0.7, colour =cc@meta.data$value)) +
+ geom_point(size = 5) +
+ scale_colour_gradient(low = "yellow", high = "blue")
By below code I obtained a tsne in link
> cols <- scales::seq_gradient_pal(low="beige", high="red", space="Lab")(seq(from=0, to=1,length.out=48))
>
> TSNEPlot(cc, colors.use=cols)
Now I want to know how I could convert this range to a 8-18, 18-28, 28-38, 38-48 colour range as a gradient of blue to yellow?
FeaturePlot(). – Kohl Kinning Oct 29 '18 at 19:41>colnames(exp)="value"? Unless I'm missing something you are trying to assign one string to a vector of values. This shouldn't work. You should be getting an error likelength of 'dimnames' [2] not equal to array extent. Do you ultimately want a plot of the values in one column of the matrix namedexp? – Kohl Kinning Oct 29 '18 at 21:44expand thematmatrices do not match. Please show a reproducible, minimal example. Also by converting the exp to a data.frame it might convert your numeric values to factors, which is why you see them with each factor into a different color. – llrs Oct 30 '18 at 10:36expis a binarized version ofmatas Feresh Teh states above.Feresh Teh: I would think you would want
– Kohl Kinning Oct 30 '18 at 14:22rowSums(), that way all of the columns for a given row will be reduced to the sum and you'll have a value (expression?) for each cell. When you add metadata to a Seurat object, it will be in this format--a value for each cell.expisexp <- mat;exp[exp>2] <- 1;exp[exp<2] <- 0But they don't match the row names between the twoheads shown. – llrs Oct 30 '18 at 14:32colSums()makes sense. The cell names are thes1.1, s1.2etc. Working on an answer. – Kohl Kinning Oct 30 '18 at 18:57