I would like to make a 3D PCA but not sure how to label group wise which i can do for 2D PCA 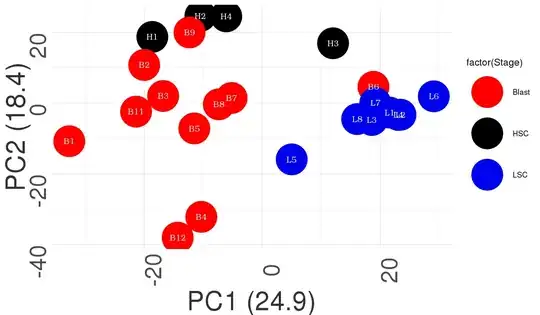
library(ggplot2)
test<- read.csv("NEW_RBP/TF_DISEASE.txt",header = T,row.names = 1,sep = '\t')
Sample = c(rep("HSC",4),rep("Blast",11),rep("LSC",8))
Stage=c(rep("HSC",4),rep("Blast",11),rep("LSC",8))
df_final=data.frame(Sample,Stage)
df_final
pca_data=prcomp(t(test), center=TRUE, scale=TRUE)
pca_data_perc=round(100*pca_data$sdev^2/sum(pca_data$sdev^2),1)
df_pca_data=data.frame(PC1 = pca_data$x[,1],
PC2 = pca_data$x[,2], sample = colnames(test))
myColors <- c("red", "black", "blue2")
sample <- as.vector(df_pca_data$sample)
ggplot(df_pca_data, aes(PC1,PC2, colour = factor(Stage)))+#,label=rownames(t(test)))+
geom_point(size=30)+
geom_text(aes(label=sample),color="white",size=6,angle =0,parse = TRUE, family="Bookman", fontface="bold")+
labs(x=paste0("PC1 (",pca_data_perc[1],")"), y=paste0("PC2 (",pca_data_perc[2],")"))+
theme_minimal(base_size=20) +
theme(axis.text.x=element_text(size=rel(3), angle=90))+
theme(axis.text.y=element_text(size=rel(3), angle=90))+
theme(axis.title.x = element_text(colour="grey20",size=55,angle=0,hjust=.5,vjust=0,face="plain"))+
theme(axis.title.y = element_text(colour="grey20",size=55,angle=90,hjust=.5,vjust=.5,face="plain"))+
scale_color_manual(values=myColors)
The above code give me the figure as i have attached
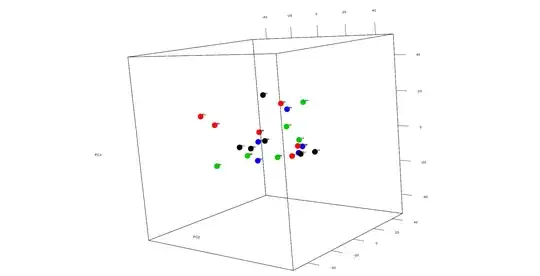
For 3D PCA I tried this
scores = as.data.frame(pca_data$x)
head(scores)
plot3d(scores[,1:3], col=c(1:4), size=20, type='p',
xlim = c(-50,50), ylim=c(-50,50), zlim=c(-50,50))
text3d(scores[,1]+2, scores[,2]+1, scores[,3]+1,
texts=c(rownames(scores)), cex= 0.7, pos=3)
I get something like this as in case of 2D PCA i can label lets say all of the HSC sample as black and so on..Im not sure how to do that for 3D as I tried using factor i couldn;t do
Any help or suggestion would be highly appreciated
cbind()andrep()functions in R – Ammar Sabir Cheema Oct 31 '18 at 15:44