Perhaps, grep is not the best tool to use in this case, but it should be in principle possible by using grep & sed. Here is an example showing three symbols around a match.
zcat My_Hiseq_Data.fq.gz | \
grep -Eo '.{0,3}GATCGATC.*' | \
sed -En 's/.*/ \0/; s/.*(.{3}GATCGATC.{0,3}).*/\1/p' | \
grep --color=always GATCGATC
Here are some explanations:
zcat will just decompress a file.grep will output all the matches with 3-symbol context (or less). This is needed because a line can have multiple matches.- first
sed command will add 3 symbols before all the matches.
- second
sed command will output the sequence with 3 surrounding characters on the left and max 3 characters on the right. This is an equivalent of grep -Eo '.{3}GATCGATC.{0,3}'.
- Finally, now that all matches are aligned,
grep will color your sequence.
This code is possible to adapt to include more symbols of context.
I would recommend doing this in another language, though. E.g., in Python:
#!/usr/bin/env python3
import sys
import re
HL = '\033[93m'
ENDC = '\033[0m'
filename = sys.argv[1]
seq = sys.argv[2]
context = 10
seqre = re.compile(r'(.{0,%d})(%s)(.{0,%d})' % (context, sys.argv[2], context))
for line in open(filename):
line = line.rstrip()
for m in seqre.finditer(line):
print(m.group(1).rjust(context),
HL,
m.group(2),
ENDC,
m.group(3),
sep='')
UPD
A more advanced Python version inspired by this answer with support for multiple files, gzip, etc.
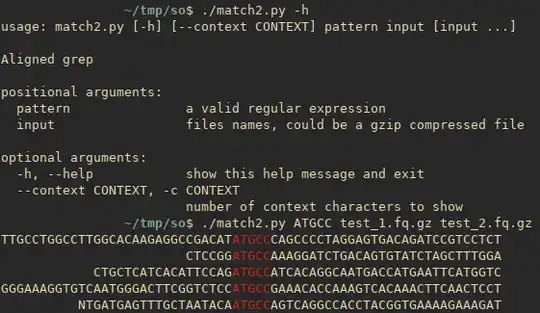
#!/usr/bin/env python3
import sys
import re
import argparse
import gzip
from termcolor import colored
def align_matches(f, seqre, context):
# for every line
for line in f:
# get rid of newline character
line = line.rstrip()
for m in seqre.finditer(line):
# for every match print right-aligned left-context,
# match colored in red, and right-context
print(m.group(1).rjust(context),
colored(m.group(2), 'red'),
m.group(3),
sep='')
# this function will check file extension to be able
# to open text files as well as gzip-compressed files
def myopen(fn):
if fn.endswith('.gz'):
return gzip.open(fn, 'rt')
else:
return open(fn, 'rt')
if __name__ == '__main__':
# define script arguments
parser = argparse.ArgumentParser(description='Aligned grep')
parser.add_argument('pattern', type=str,
help='a valid regular expression')
parser.add_argument('input', type=myopen, nargs='+',
help='files names, could be a gzip compressed file ending with .gz')
parser.add_argument('--context', '-c', default=30, type=int,
help='number of context characters to show')
args = parser.parse_args()
# compile a regular expression with a match surrounded
# by at most args.context number of characters
seqre = re.compile(r'(.{0,%d})(%s)(.{0,%d})' % (
args.context, args.pattern, args.context))
# for every input file run align_matches
for f in args.input:
align_matches(f, seqre, args.context)



$left = " " x ($context - length($left)) if length($left) < $context; – Peter Menzel Oct 28 '18 at 11:02