I am trying to find details for how the ENCODE ChIP seq samples were processed.
Linking to a ChIPseq samples on www.encodeproject.org (e.g. https://www.encodeproject.org/experiments/ENCSR051DXE/) offers a PDF "overview" of the pipeline which says basically nothing. The page will also offer you a map of the pipeline, which basically just has headings:
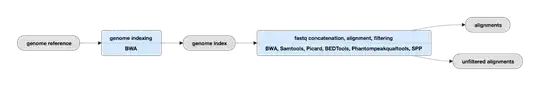
Clicking on a step gives you a popup with a link to the "Chip-seq read mapping" pipeline. This just contains the same overview PDF and a link to "ChIP-seq Data Standards and Processing Pipeline". This page contains again, a vague outline of the pipeline, a link back to the page we just came from and a list of quality control thresholds.
I can't seem to find anywhere the proper details. For example, if BWA was used, was it MEM or aln? What parameters were used for the mapping? One of the steps is read filtering: what where the filters? How were they applied? There is a link to a github page for running the pipeline on DNAnexus, but I can't make head nor tail of it.
bwa alnoption with-q 5 -l 32 -k 2. Ref: https://github.com/ENCODE-DCC/chip-seq-pipeline/blob/541dd361f28ef7568f3743c00dfcb882c461df2c/dnanexus/input_shield/dxapp.json line 43 – arup Oct 24 '18 at 12:02bwa alnRef: https://github.com/ENCODE-DCC/chip-seq-pipeline2/blob/e4d53da91f7dcf16dad984b1c9ebbbf168f8ef4b/src/encode_bwa.py line: 73. – arup Oct 24 '18 at 12:17