I am using chromPlot to visualise the genome of C. elegans.
library(chromPlot)
I have created the following data frame with the lengths of C. elegans chromosomes.
Chrom Start End Name
1 1 0 15072434 contigs
2 2 0 15279421 contigs
3 3 0 13783801 contigs
4 4 0 17493829 contigs
5 6 0 20924180 contigs
6 7 0 17718942 contigs
7 5 0 13794 contigs
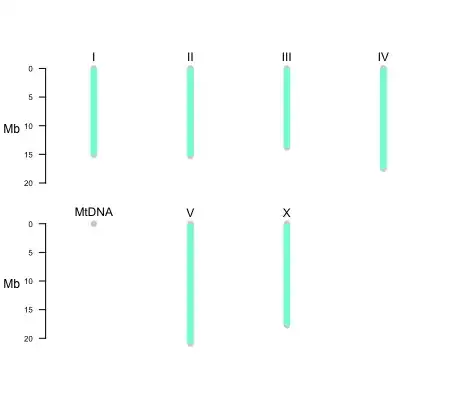
When I use chromPlot(dataframe), I get a plot with 4 chromosomes and the following output/error:
1 segment clusters.
1 segment clusters.
1 segment clusters.
1 segment clusters.
1 segment clusters.
1 segment clusters.
1 segment clusters.
Chrom 1 : 15072434 bp
Chrom 2 : 15279421 bp
Chrom 3 : 13783801 bp
Chrom 4 : 17493829 bp
Error in seq.default(minval, maxval, length = howmany) :
'from' must be of length 1
Does anybody know why I get this error?
I don't get this error when I use the data frame provided by the package with human segments. The structure of the data frame is:
'data.frame': 457 obs. of 4 variables:
$ Chrom: chr "1" "1" "1" "1" ...
$ Start: int 124535434 121535434 3845268 13219912 17125658 29878082 120697156 120936695 121485434 142731022 ...
$ End : int 142535434 124535434 3995268 13319912 17175658 30028082 120747156 121086695 121535434 142781022 ...
$ Name : chr "heterochromatin" "centromere" "contig" "contig" ...
Compared to my data frame:
'data.frame': 7 obs. of 4 variables:
$ Chrom: chr "1" "2" "3" "4" ...
$ Start: int 0 0 0 0 0 0 0
$ End : int 15072434 15279421 13783801 17493829 20924180 17718942 13794
$ Name : chr "contigs" "contigs" "contigs" "contigs" ...
I am on MacOS X.
Update
The 20th line of the package-provided hg_gap data frame is:
Chrom Start End Name
20 1 0 10000 telomere
Note that the Start is 0.
I've also tried changing the start from 0 to 1. The error stays the same.
Update 2
Sorting the data frame by chromosome name does not improve the situation.
The traceback of the error is:
6: stop("'from' must be of length 1")
5: seq.default(minval, maxval, length = howmany)
4: seq(minval, maxval, length = howmany)
3: pretty_ticks(minlab, maxlab, 2, ...)
2: draw.scale(y = 0.8 * chr.length[[chrom]] + 0.2 * xylims[3], minval = margin,
maxval = maxGeneCount, minlab = annot1_plot_range[1], maxlab = annot1_plot_range[2],
lwd = 4, col = colAnnot1, cex = cex, title = scale.title)
1: chromPlot::chromPlot(gaps)
I have emailed the maintainer of the package Karen Orostica with a link to this post.
startshould be 1 or more and not zero. I think the BED format should be 1 based (now it is 0 based). – benn Jun 27 '18 at 14:16Chromneeds to be sorted, in your example afterChrom1-4, comes 6 not 5. I am just guessing here. – benn Jun 27 '18 at 14:27seq(NULL, to = 50). If the data is valid, it is a bug on the package. Create an issue as required by the maintainer of the package adding thetracebackto see the origin of the error. – llrs Jun 27 '18 at 14:27