I am trying to estimate a phylogenetic tree in R with ape and phangorn. With pratchet() and optim.parsimony(), I can estimate a phylogenetic tree, but branch lengths are not estimated. Is there a way to estimate a phylogeny with branch lengths with the following made-up data:
x <- data.frame("L1" = c(0,0,1,0,0,0,1,1,1,1,0,0,0,0,1), "L2" = c(0,1,1,0,1,1,0,1,1,0,1,1,0,1,1),"L3" = c(1,1,1,0,0,0,1,1,1,0,0,0,1,1,1), "L4"=c(0,0,1,1,0,0,0,0,1,1,0,0,0,0,1))
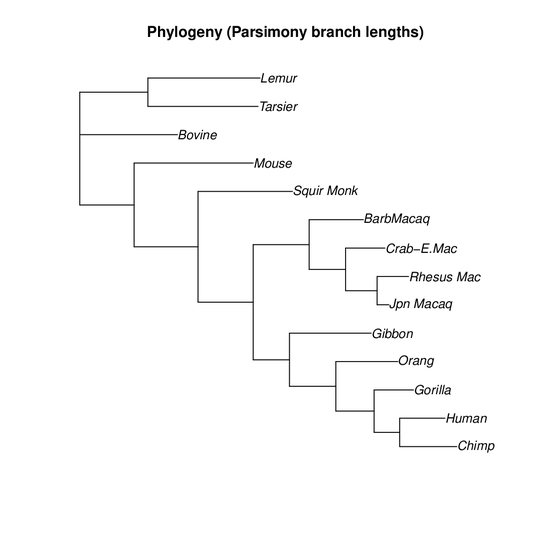
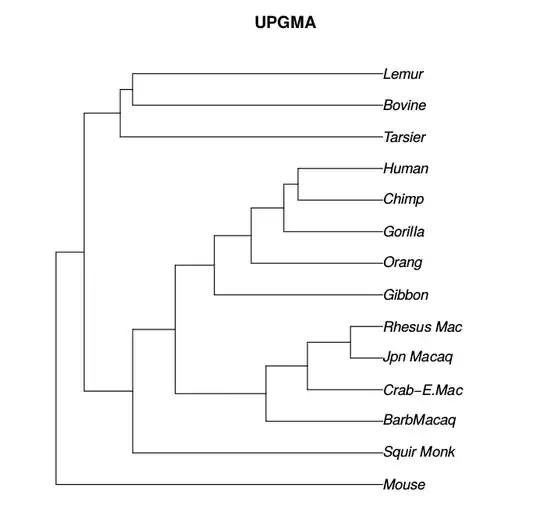
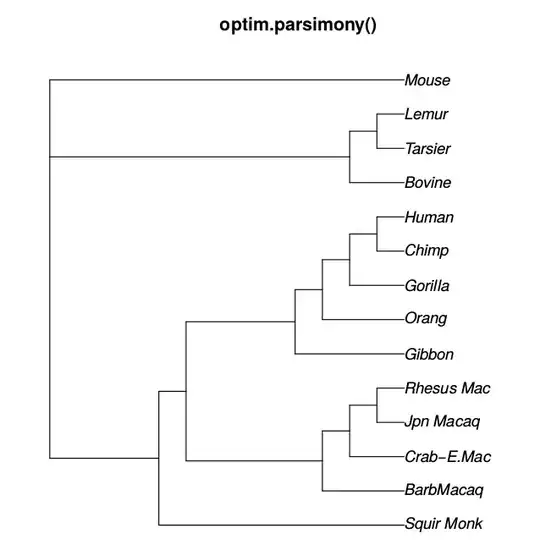
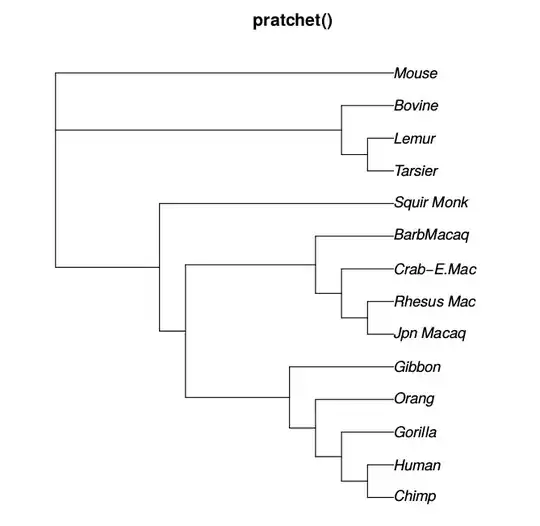
L1,L2, etc.; not"L1","L2"…. R unfortunately allows is but this is a language flaw that should not be exploited. – Konrad Rudolph Jun 26 '18 at 12:35