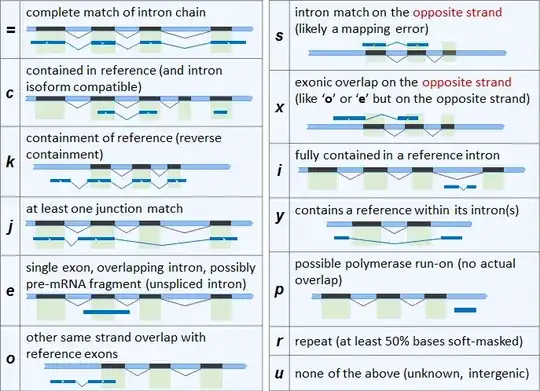
I used gffcompare recently to compare a newly assembled transcriptome and a published transcriptome. The class code "p" transcripts are "possible polymerase run on".
What does that mean? I tried very hard but still cannot find the answer.
I used gffcompare recently to compare a newly assembled transcriptome and a published transcriptome. The class code "p" transcripts are "possible polymerase run on".

What does that mean? I tried very hard but still cannot find the answer.
On most coding genes (with the exception of replication dependent histone genes), Transcripts (as opposed to transcription) are terminated by cleavage polyadenylation sites, where the growing transcript is cleaved by a protein complex and the poly-A tail. However, PolII continues to transcribe until it is terminated by torpedo termination (see here). Normally this extra RNA after the polyA site is degraded by Xrn2, but if you capture any particular snapshot of the RNA in the cell at any one time there will be a small amount that corresponds to this run on transcription.
The general idea behind a run-on fragment is that it's background noise. This derives from an open area of the genome that's next to an area that's actually being transcribed. Thus, all of the machinery for transcription is already present, so if the region adopts a bit more of an open conformation then that machinery can sporadically bind and start transcribing. You can get those either up- or down-stream of genes. A lot of things like this are just background transcriptional noise.