I am working on haplotype data and want to make a tree out of haplogroups using ggtree. I have following data in newick format.
(B(B2b_M112),C(C_RPS4Y711,C1a_V20,C2_M217,C2b1a_F3985,C2C1_Z1338),E(E1b1a_V38,E1b1b_M215,E2_M75),G(G_M201,G1_M342,G2a1_Z6552,G2a2a_PF3147,G2a2b2b_PF3359,G2a2b1_M406,G2a2b2a1a_U1,G2a2b2a1b_L497,G2a2b2a1c_CTS342,G2a_L30,G2b_M3155,G2b_M283),H(H1_M69,H1a_M82),I(I_M170,I1_M253,I2a1_Isles,I2a1_S21825,I2a1a_M26,I2a1b3_L621,I2a2a_M223,I2a2b_L38,I2c2_Y16419),J(J_M304,J1a_PF7257,J1a2a1a2_P58,J1a2a2_PF7264,J1a3_Z1828,J2_M172,J2a_L25,J2a1_L26,J2a1_M319,J2a1_Z1846,J2a1_Z387,J2a1_Z467,J2a1_Z6046,J2a1_Z6063,J2a1_Z6065,J2a1_Z7671,J2a1_Z7700,J2a2_PF5008, J2b_M241,J2b1_M205,J2b2_M241,J2b2_Z2456),K(K_M9),L(L_M20,L1_M22,L1a_M27,L1b_M317,L1c_M357),N(N3,N1a1_M46,N1a2_L666),O(O1a1_B384,O1b1_F2320,O1b2_P49,O2 IMS_JST0213554,O2a1_F51,O_O2a2_F525,O3_M122),Q(Q F1096,Q_L27,Q_L56,Q_M24,Q_M346),R(R_M207,M734,P224,P280xM173,R1a_M417,R1a1_M459,R1a1a_M17,R1b_M343,R1b_M73,R1b1a1b_V1636,R1b1a2_V88,R1b1b_PH155,R2_M479,R2a_M124),T(T_M184,T_L131,T_PF5633));
I am using following R code to plot a tree from this data,
> library("ggtree")
> tree <- read.tree(text = "(B(B2b_M112),C(C_RPS4Y711,C1a_V20,C2_M217,C2b1a_F3985,C2C1_Z1338),E(E1b1a_V38,E1b1b_M215,E2_M75),G(G_M201,G1_M342,G2a1_Z6552,G2a2a_PF3147,G2a2b2b_PF3359,G2a2b1_M406,G2a2b2a1a_U1,G2a2b2a1b_L497,G2a2b2a1c_CTS342,G2a_L30,G2b_M3155,G2b_M283),H(H1_M69,H1a_M82),I(I_M170,I1_M253,I2a1_Isles,I2a1_S21825,I2a1a_M26,I2a1b3_L621,I2a2a_M223,I2a2b_L38,I2c2_Y16419),J(J_M304,J1a_PF7257,J1a2a1a2_P58,J1a2a2_PF7264,J1a3_Z1828,J2_M172,J2a_L25,J2a1_L26,J2a1_M319,J2a1_Z1846,J2a1_Z387,J2a1_Z467,J2a1_Z6046,J2a1_Z6063,J2a1_Z6065,J2a1_Z7671,J2a1_Z7700,J2a2_PF5008, J2b_M241,J2b1_M205,J2b2_M241,J2b2_Z2456),K(K_M9),L(L_M20,L1_M22,L1a_M27,L1b_M317,L1c_M357),N(N3,N1a1_M46,N1a2_L666),O(O1a1_B384,O1b1_F2320,O1b2_P49,O2 IMS_JST0213554,O2a1_F51,O_O2a2_F525,O3_M122),Q(Q F1096,Q_L27,Q_L56,Q_M24,Q_M346),R(R_M207,M734,P224,P280xM173,R1a_M417,R1a1_M459,R1a1a_M17,R1b_M343,R1b_M73,R1b1a1b_V1636,R1b1a2_V88,R1b1b_PH155,R2_M479,R2a_M124),T(T_M184,T_L131,T_PF5633));")
> p<-ggtree(tree)
> p+geom_tiplab()
And the displayed tree looks like this

I have following problems in this tree,
- It gives a low quality image
- It does not show the haplotype taxa names clearly
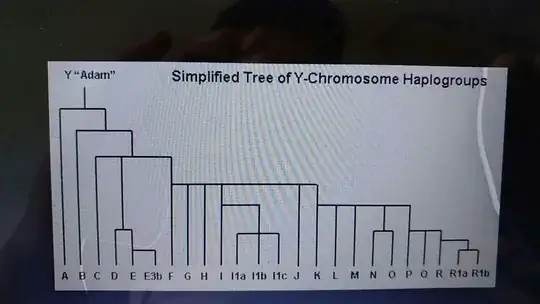
- I want to arrange haplogroups i.e nodes in the desired according to the tree given below,
i.e B haplogroups with C haplotypes and with C, E named haplotypes etc.
I have tried to solve these problems but failed to find any solution. Is there a solution?
tiplabissue, pls refer to the FAQ. – Guangchuang Yu May 28 '18 at 02:20