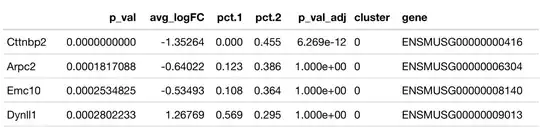
I compared two manually defined clusters using Seurat package function FindAllMarkers and got the output:
Now, I am confused about three things:
- What are
pct.1andpct.2? - How come
p-adjusted valuesequal to1? What does it mean? - If we take first row, what does
avg_logFCvalue of-1.35264mean when we have cluster0in theclustercolumn? Is it that in cluster0theCttnbp2gene downregulated by a factor of2^1.35264?