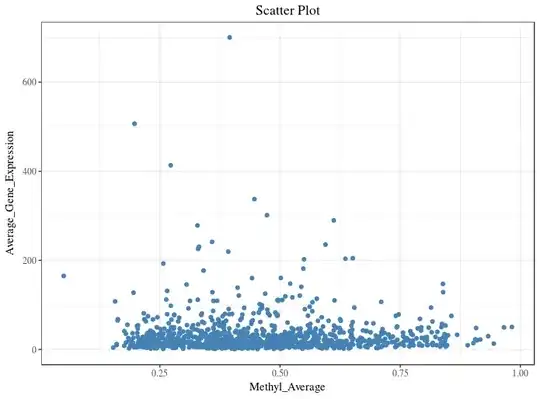
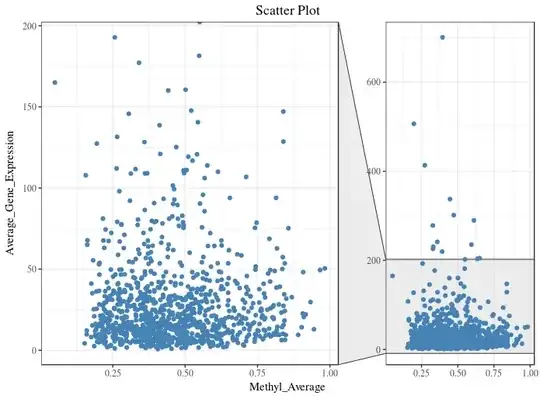
I am using ggplot2 to plot a scatter plot.
library(ggforce) # required by facet_zoom
g <- ggplot(data.plot, aes(x = Methyl_Average, y = Average_Gene_Expression))
g1 <- g + geom_point(color = "steelblue") + theme_bw(base_family = "Times") +
ggtitle( "Scatter Plot") +
theme_bw(base_family = "Times") +
theme(plot.title = element_text(hjust = 0.5))
As most of my data points are in the range of 0 - 200 on y-axis, I want to zoom this section. I tried doing it using facet_zoom from ggforce package :
g1 + facet_zoom(y = Average_Gene_Expression > 0 & Average_Gene_Expression < 200)
Instead of two plots in one figure, I want one plot (like first figure), where 80% of y-axis displays the data points in the range of 0 - 200 and 20% of the y-axis displays the remaining and x-axis unaltered. Which function/package should I use do it in R?