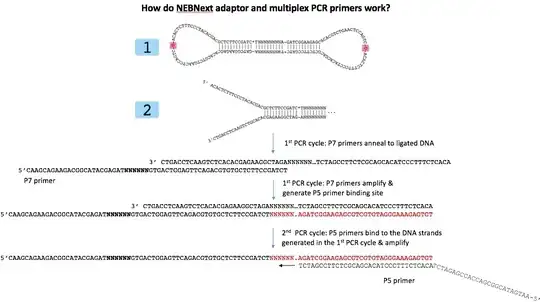
I am making some infographics of different library prep types and noticed something weird about NEB's Ultra II adapters.
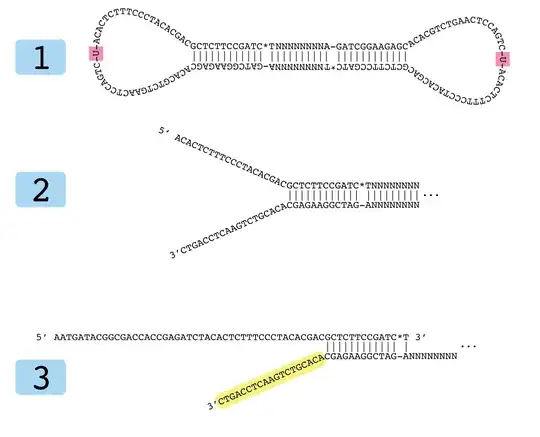
Molecule 1 is the desired product of the adapter ligation step. * indicates a phosphorothioate bond and - indicates a normal bond. | indicates complementary sequence through hydrogen bonding.
Molecule 2 is the product after treatment with the USER enzyme.
Molecule 3 is the result of melting molecule 2 and hybridizing the NEBNext Universal PCR Primer for Illumina.
My question is: Why does Molecule 3 have a large non-complementary region on the 3' end of the original molecule? Is that just to force Molecule 1 to have an open loop conformation so that the USER enzyme can access it better? Or maybe it is something to do with PCR kinetics?
I understand that PCR synthesizes in the 5'->3' direction, so Molecule 3 only forms with the original adapter-ligated molecules before PCR.