Also posted on biostars.
I am trying to use cellranger or bcl2fastq to convert the .bcl files that I got from single cell analysis run into fastq files for further analysis. I needed to generate sample_sheet.csv and so I used the following tool:
which I copy-pasted into a text file using vim. sample_sheet.csv looks like that:
[Header]
EMFileVersion,4
[Reads]
26
8
98
[Data]
Lane,Sample_ID,Sample_Name,index,Sample_Project
1,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180406
1,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180406
1,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180406
1,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180406
1,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180406
1,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180406
1,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180406
1,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180406
2,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180406
2,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180406
2,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180406
2,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180406
2,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180406
2,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180406
2,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180406
2,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180406
3,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180406
3,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180406
3,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180406
3,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180406
3,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180406
3,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180406
3,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180406
3,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180406
4,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180406
4,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180406
4,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180406
4,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180406
4,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180406
4,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180406
4,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180406
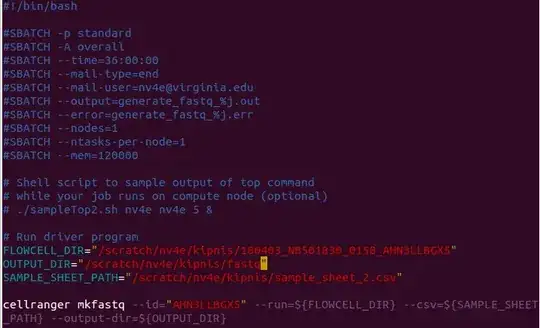
Then I ran cellranger using bash script:
#!/bin/bash
FLOWCELL_DIR="/scratch/nv4e/kipnis/180403_NB501830_0158_AHN3LLBGX5"
OUTPUT_DIR="/scratch/nv4e/kipnis/fastq"
SAMPLE_SHEET_PATH="/scratch/nv4e/kipnis/sample_sheet_2.csv"
cellranger mkfastq --id="AHN3LLBGX5" --run=${FLOWCELL_DIR} --csv=${SAMPLE_SHEET_PATH} --output-dir=${OUTPUT_DIR}
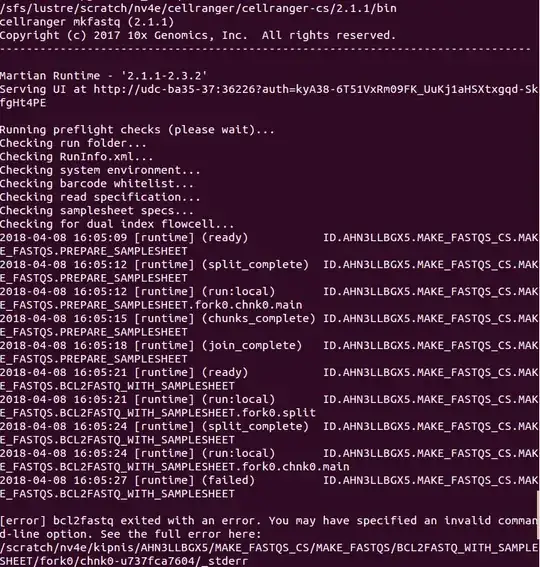
It fails with the following output:
_stderr file shows:
and that my sample_sheet.csv can not be parsed:
It could be because CLRF needs to be converted into LF, but I tried two commands to do this unsuccessfully:
tr -d '\r' < input > output and perl -pi -e 's/\r\n/\n/g' input
From the following thread:
https://unix.stackexchange.com/questions/277217/how-to-install-dos2unix-on-linux-without-root-access
The above commands successfully strip the \r from the line endings, but the error from bcl2fastq remains.
What am I doing wrong? Any suggestions would be greatly appreciated.
Update
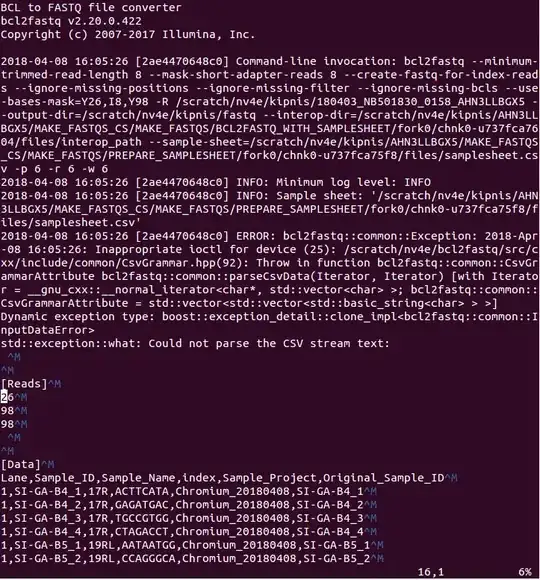
I looked into the folder that is generated when the cellranger runs and found two .csv files generated by the cellranger. One of them, samplesheet.csv, contains these ^M characters that are shown in the error:
Update
Maybe one of the issues was that I was submitting regular .sh file on the SLURM cluster. However, I wrote the right .slurm file and submitted it yesterday with sbatch command:
However, today I got the message in my email:
And neither _stderr, nor generate_fastq_id.err or generate_fastq_id.out files contain anything. I thought initially that I set not enough time and it timed out, however, it is not true: I set the time to 24h and it has been running for just 13h before failing.
Update
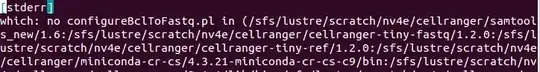
I searched over all of the files in the generated folder with the find linux command and actually found the error files that show the same error as before: issue with the sample_sheet.csv file. This error file is AHN3LLBGX5/MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/chnk0-u179acba3d8/_stderr.
So, right now the issue seems to be the following: I am feeding in the right sample_sheet.csv without the ^M symbols, but cellranger transforms it into one that contains ^M and then uses it giving the error. Above I see also which: no configureBclToFastq.pl and ERROR: bcl2fastq::common::Exception: 2018-Apr-09 13:33:14: Inappropriate ioctl for device, so maybe my input parameters are just wrong to cellranger and it has nothing to do with the sample_sheet.csv?
Update
I tried to use --sample-sheet instead of --csv since I am feeding in complex sample sheet. No luck.
Update
Here you can find all the error document in text:
which: no configureBclToFastq.pl in (/sfs/lustre/scratch/nv4e/cellranger/samtools_new/1.6:/sfs/lustre/scratch/nv4e/cellranger/cellranger-tiny-fastq/1.2.0:/sfs/lustre/scratch/nv4e/cellranger/cellranger-tiny-ref/1.2.0:/sfs/lustre/scratch/nv4e/cellranger/miniconda-cr-cs/4.3.21-miniconda-cr-cs-c9/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/lib/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/tenkit/lib/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/tenkit/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/bin:/sfs/lustre/scratch/nv4e/cellranger/lz4/v1.8.0:/sfs/lustre/scratch/nv4e/cellranger/martian-cs/2.3.2/bin:/sfs/lustre/scratch/nv4e/cellranger/STAR/5dda596:/sfs/lustre/scratch/nv4e/cellranger/samtools_new/1.6:/sfs/lustre/scratch/nv4e/cellranger/cellranger-tiny-fastq/1.2.0:/sfs/lustre/scratch/nv4e/cellranger/cellranger-tiny-ref/1.2.0:/sfs/lustre/scratch/nv4e/cellranger/miniconda-cr-cs/4.3.21-miniconda-cr-cs-c9/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/lib/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/tenkit/lib/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/tenkit/bin:/sfs/lustre/scratch/nv4e/cellranger/cellranger-cs/2.1.1/bin:/sfs/lustre/scratch/nv4e/cellranger/lz4/v1.8.0:/sfs/lustre/scratch/nv4e/cellranger/martian-cs/2.3.2/bin:/sfs/lustre/scratch/nv4e/cellranger/STAR/5dda596:/scratch/nv4e/cellranger:/scratch/nv4e/bcl2fastq/build/bin:/scratch/nv4e/spark/bin:/scratch/nv4e/scala/bin:/sfs/nfs/blue/nv4e/private/bin:/sfs/nfs/blue/nv4e/anaconda2/bin:/sfs/nfs/blue/nv4e/.local/bin:/usr/lib64/qt-3.3/bin:/usr/local/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/opt/slurm/current/bin:/opt/slurm/current/sbin:/opt/singularity/current/bin:/opt/rci/bin:/opt/rci/sbin:/opt/nhc/current/sbin:/share/rci_apps/common/bin:/share/resources/HPCtools/) BCL to FASTQ file converter bcl2fastq v2.20.0.422 Copyright (c) 2007-2017 Illumina, Inc. 2018-04-09 18:31:29 [7fcdb39f37c0] Command-line invocation: bcl2fastq --minimum-trimmed-read-length 8 --mask-short-adapter-reads 8 --create-fastq-for-index-reads --ignore-missing-positions --ignore-missing-filter --ignore-missing-bcls --use-bases-mask=Y26,I8,Y98 -R /scratch/nv4e/kipnis/180403_NB501830_0158_AHN3LLBGX5 --output-dir=/scratch/nv4e/kipnis/AHN3LLBGX5/MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/chnk0-u21f1cbe9bf/files/fastq_path --interop-dir=/scratch/nv4e/kipnis/AHN3LLBGX5/MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/chnk0-u21f1cbe9bf/files/interop_path --sample-sheet=/scratch/nv4e/kipnis/AHN3LLBGX5/MAKE_FASTQS_CS/MAKE_FASTQS/PREPARE_SAMPLESHEET/fork0/chnk0-u09e7cbe891/files/samplesheet.csv -p 6 -r 6 -w 6 2018-04-09 18:31:29 [7fcdb39f37c0] INFO: Minimum log level: INFO 2018-04-09 18:31:29 [7fcdb39f37c0] INFO: Sample sheet: '/scratch/nv4e/kipnis/AHN3LLBGX5/MAKE_FASTQS_CS/MAKE_FASTQS/PREPARE_SAMPLESHEET/fork0/chnk0-u09e7cbe891/files/samplesheet.csv' 2018-04-09 18:31:29 [7fcdb39f37c0] ERROR: bcl2fastq::common::Exception: 2018-Apr-09 18:31:29: Inappropriate ioctl for device (25): /scratch/nv4e/bcl2fastq/src/cxx/include/common/CsvGrammar.hpp(92): Throw in function bcl2fastq::common::CsvGrammarAttribute bcl2fastq::common::parseCsvData(Iterator, Iterator) [with Iterator = __gnu_cxx::__normal_iterator >; bcl2fastq::common::CsvGrammarAttribute = std::vector > >] Dynamic exception type: boost::exception_detail::clone_impl std::exception::what: Could not parse the CSV stream text: ^M ^M [Reads]^M 26^M 98^M ^M ^M [Data]^M Lane,Sample_ID,Sample_Name,index,Sample_Project,Original_Sample_ID^M 1,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180409,SI-GA-B4_1^M 1,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180409,SI-GA-B4_2^M 1,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180409,SI-GA-B4_3^M 1,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180409,SI-GA-B4_4^M 1,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180409,SI-GA-B5_1^M 1,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180409,SI-GA-B5_2^M 1,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180409,SI-GA-B5_3^M 1,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180409,SI-GA-B5_4^M 2,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180409,SI-GA-B4_1^M 2,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180409,SI-GA-B4_2^M 2,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180409,SI-GA-B4_3^M 2,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180409,SI-GA-B4_4^M 2,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180409,SI-GA-B5_1^M 2,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180409,SI-GA-B5_2^M 2,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180409,SI-GA-B5_3^M 2,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180409,SI-GA-B5_4^M 3,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180409,SI-GA-B4_1^M 3,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180409,SI-GA-B4_2^M 3,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180409,SI-GA-B4_3^M 3,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180409,SI-GA-B4_4^M 3,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180409,SI-GA-B5_1^M 3,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180409,SI-GA-B5_2^M 3,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180409,SI-GA-B5_3^M 3,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180409,SI-GA-B5_4^M 4,SI-GA-B4_1,17R,ACTTCATA,Chromium_20180409,SI-GA-B4_1^M 4,SI-GA-B4_2,17R,GAGATGAC,Chromium_20180409,SI-GA-B4_2^M 4,SI-GA-B4_3,17R,TGCCGTGG,Chromium_20180409,SI-GA-B4_3^M 4,SI-GA-B4_4,17R,CTAGACCT,Chromium_20180409,SI-GA-B4_4^M 4,SI-GA-B5_1,19RL,AATAATGG,Chromium_20180409,SI-GA-B5_1^M 4,SI-GA-B5_2,19RL,CCAGGGCA,Chromium_20180409,SI-GA-B5_2^M 4,SI-GA-B5_3,19RL,TGCCTCAT,Chromium_20180409,SI-GA-B5_3^M 4,SI-GA-B5_4,19RL,GTGTCATC,Chromium_20180409,SI-GA-B5_4^M

_stderrfile they are somehow present – Nikita Vlasenko Apr 08 '18 at 23:45_stderrafter removing\r? Because the^Mcharacters indicate a line break problem. If they go away after running thetr/perlcommand, then there's probably an additional problem – heathobrien Apr 09 '18 at 09:00bcl2fastqwith your sample sheet (i.e., without using cellranger)? – Devon Ryan Apr 10 '18 at 19:17