I am calling variants from a human sample using bwa mem to align the reads and gatk to call the variants. I'm trying to understand why a specific variant was not called in my sample. I have checked the bam alignments in a GUI viewer and I can see that there are reads supporting the missing variant. It looks like the issue is a low allelic balance, with far more reads supporting the reference than the alternate allele but I want to get the actual numbers.
So, given a specific variant like this:
chr22 425236 C T
How can I count the number of reads in my sample.bam file that support that variant and the number that don't on Linux?
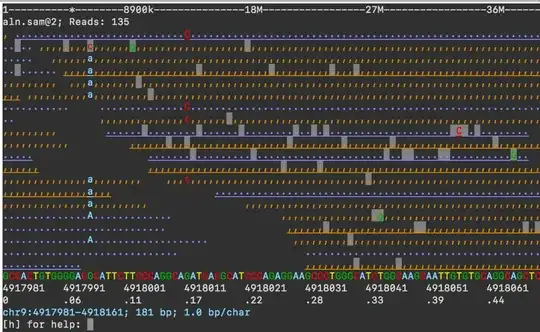
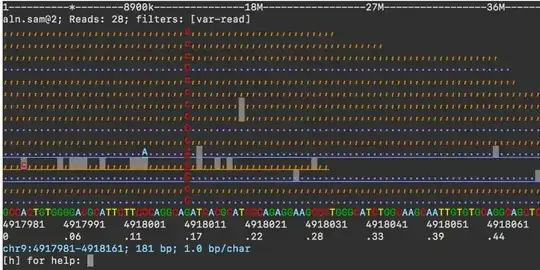
conda install asciigenomeshould also work. Please submit an issue to GitHub or post a question with more detail. – dariober Jun 10 '21 at 07:44