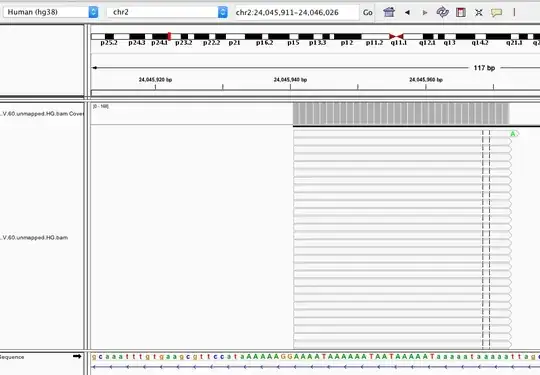
I have a screenshot where many of my reads were aligned to a region that I suspect a low complexity region. Although you can't see, all those reads are clipped in the cigar strings. Sample cigar strings: 64S32M29S, 74S32M18S etc... Consequently, the actual sequence mapped to the genome is less than the read length.
I have a feeling that my alignments are bogus because of the complexity and clipping, but I'm not sure if the complexity is indeed low. All I see is a few "T" between a bunch of "A". How to define a low complexity region?
Q: Is this a low complexity region in the human hg38 genome? Would that be fair if I report "the alignments are likely be errors due to the low complexity and alignment clipping"?