I am preprocessing scRNA-seq data. What is the best practice in use to run both ComBat for batch effects removal, data imputation (to mitigate dropout) and library size normalization?
I thought that library size should be run first, since it is per-cell normalization, then ComBat batch effects removal. On the original paper - Johnson et al. (2007) - it is stated that:
We assume that the data have been normalized and expression values have been estimated for all genes and samples.
However, I want to apply it to scRNA-seq data. Does this statement still hold? Additionally, I plan to apply imputation (e.g. with MAGIC) in the end. Is there any problem you can spot?
Update
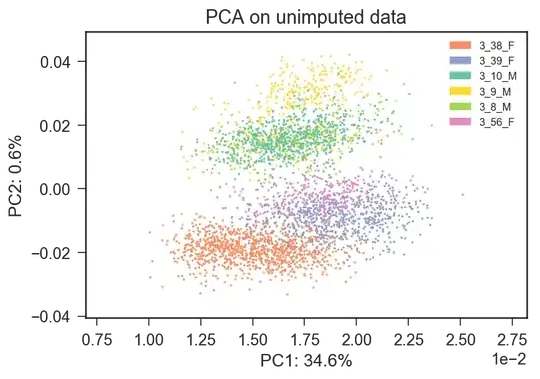
I attach the PCA regarding an example Mus Musculus dataset in which different colors represent different mice. It seems clear to me that the first two principal components are affected by batches (mouse id).
Update 2
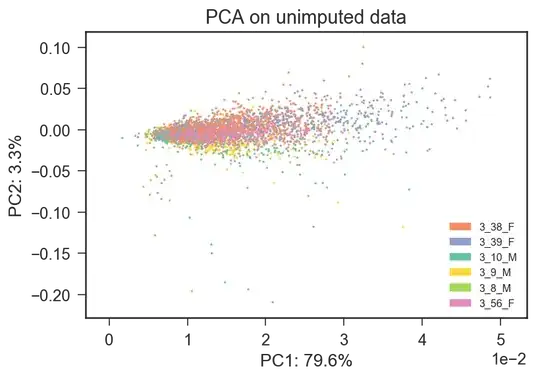
I rerun the PCA on raw counts data (the first PCA was on log-transformed data) and I obtain a different description of the dataset, in which batch effects seem not to be prevalent.