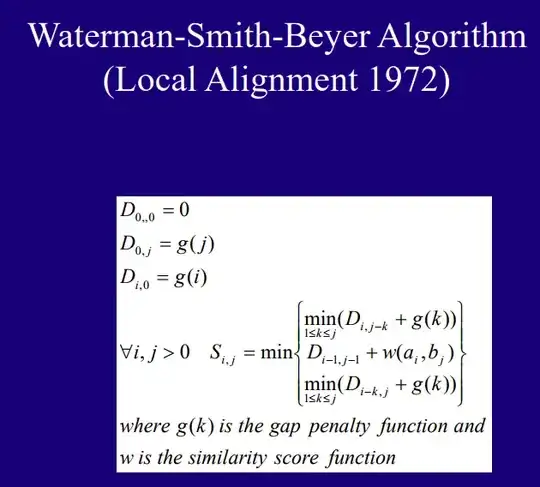
The linear gap penalty has a cost which is directly proportional to the length of the gap: the gap costs 1 unit per consecutive missing base.
The simplest case is assigning each unit (= each missing base) a penalty of 1:
g(k) = k
Or more generally, if you want to increase the penalty per base to 2, 3 or n:
g(k) = n * k
with k = length of the gap, and n = cost of 1 missing base
The affine gap penalty has a cost that follows -as its name conveniently states- an affine function.
The idea behind it, is that any gap in an alignment has a cost with two components:
- the "gap opening" penalty: it is a cost payed only once per gap, basically just for existing.
- the "gap extension" penalty: equivalent to the linear gap penalty, it costs 1 unit for each consecutive missing base.
The general formula for the cost is then:
g(k) = a + n*k
with a= opening penalty, n= extension penalty, k= length of the gap
Compared to the linear gap penalty, this will give a small bonus to longer consecutive gaps compared to skipping one nucleotide, matching one, skipping one again etc (because in the case of longer gaps, you only pay the opening penalty once, so the global cost per base decreases).
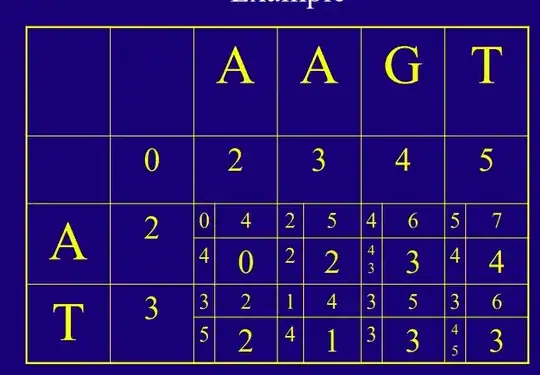
Cells on your diagram:
From what I understand, you're working with an affine penalty of g(k) = 1 + 1*k. For the [3,4] you're "coming" from the score of 2 on the left (cell 1,2). You either:
- match the A (row 1) with G (column 3) and get a penalty of 1 for substitution -> total of previous 2 + new 1 = 3
- open a gap to skip the G (column 3) and get a penalty of 2 (1 for opening + 1 because gap length of 1) -> total of previous 2 + new 2 = 4.
So you choose the lesser penalty and go with 3, which is the number selected in the bigger box.
The explanation is the exact same one for the other cell you mentioned.