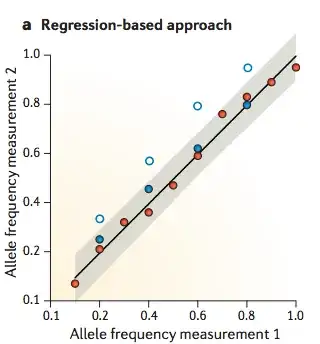
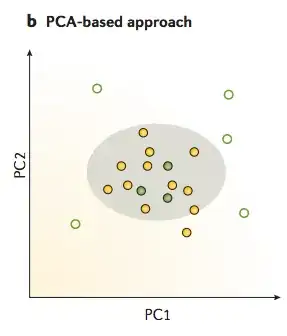
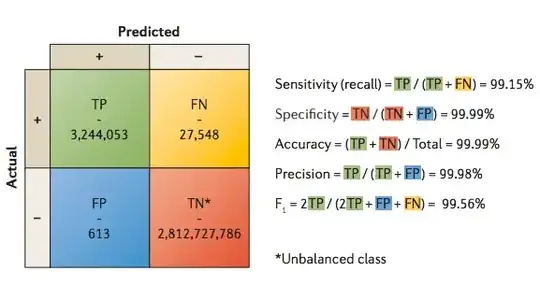
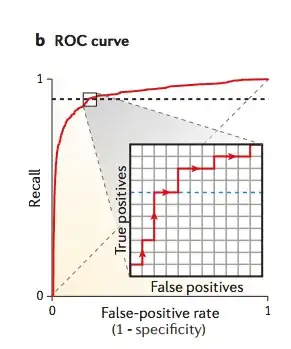
I would like to know what do people verify when designing/using spike-in controls, to be used in sequencing experiments (mainly Illumina). So far I came up with this list:
- Does it align only to a given genome synthetic reference?
- Does it contain G-quadruplexes? E.g. QGRS Mapper
- Does it contain known Illumina error motifs? (Do this matter anymore for recent platforms/chemistries?) See this article
- Does it contain other motifs known to be polymerase inhibitors? Ref
Anything else?