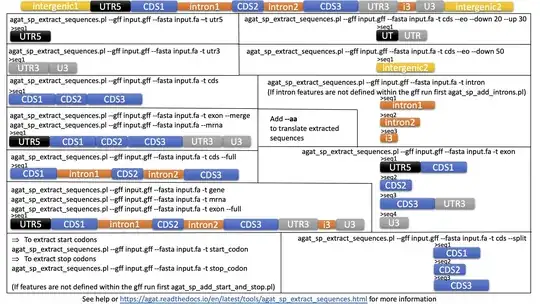
I would like to extract all the CDS from a batch of genomes. I have found a perl script from BioStars but this does not seem to work for me. I would preferably like to have a script/ method which will use the locus tag as a header.
Example
My gff files are from prokka output
ATCC0000 Prodigal:2.6 CDS 243 434 . + 0 ID=FKKLIMLP_00001;Parent=FKKLIMLP_00001_gene;inference=ab initio prediction:Prodigal:2.6;locus_tag=FKKLIMLP_00001;product=hypothetical protein
ATCC0000 prokka gene 243 434 . + . ID=FKKLIMLP_00001_gene;locus_tag=FKKLIMLP_00001
ATCC0000 Prodigal:2.6 CDS 1727 2131 . - 0 ID=FKKLIMLP_00002;Parent=FKKLIMLP_00002_gene;inference=ab initio prediction:Prodigal:2.6;locus_tag=FKKLIMLP_00002;product=hypothetical protein